Common Evolutionary Origin of Procapsid Proteases, Phage Tail Tubes, and Tubes of Bacterial Type VI Secretion Systems.
Fokine, A., Rossmann, M.G.(2016) Structure 24: 1928-1935
- PubMed: 27667692
- DOI: https://doi.org/10.1016/j.str.2016.08.013
- Primary Citation of Related Structures:
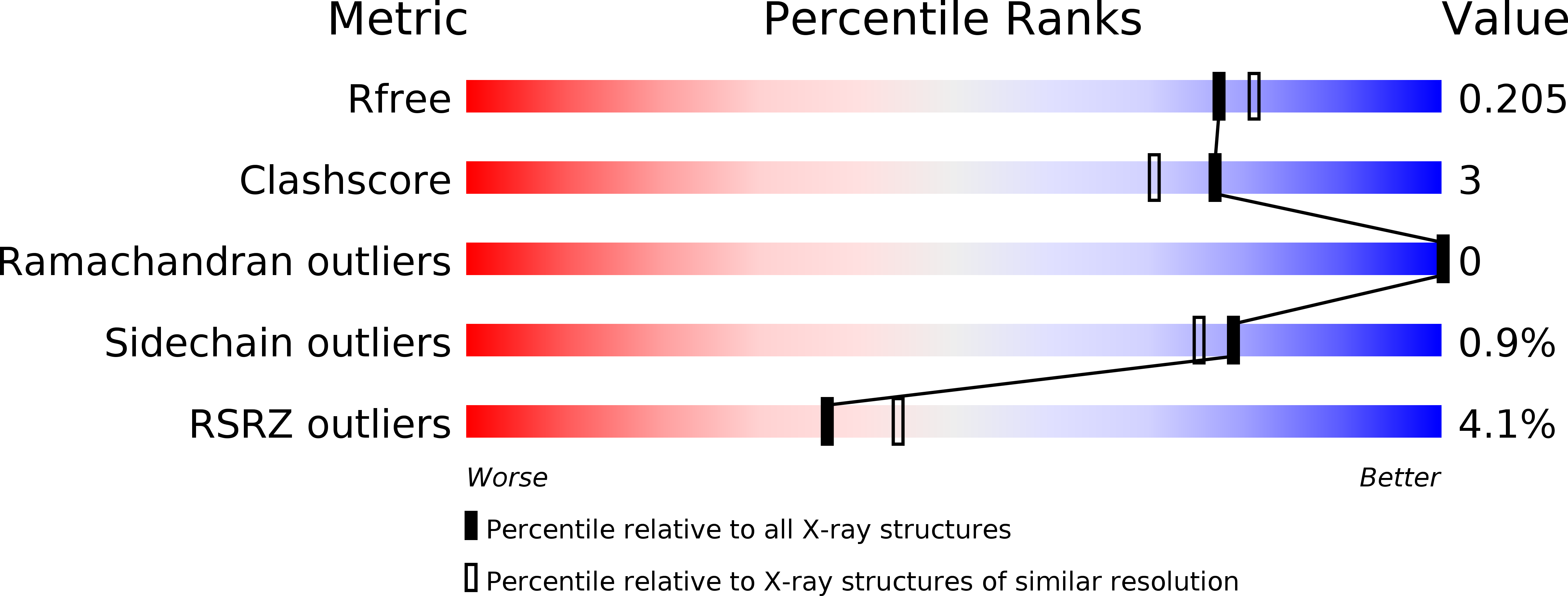
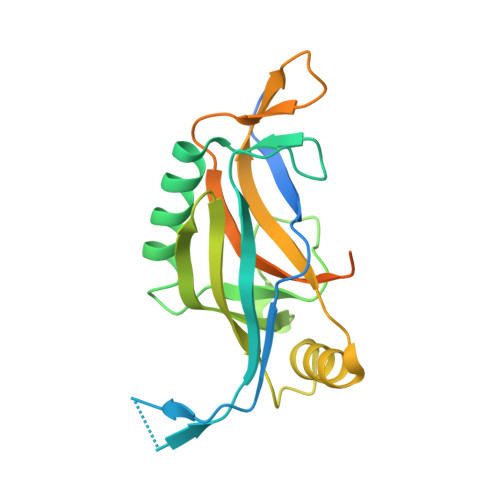
5JBL - PubMed Abstract:
Many large viruses, including tailed dsDNA bacteriophages and herpesviruses, assemble their capsids via formation of precursors, called procapsids or proheads. The prohead has an internal core, made of scaffolding proteins, and an outer shell, formed by the major capsid protein. The prohead usually contains a protease, which is activated during capsid maturation to destroy the inner core and liberate space for the genome. Here, we report a 2.0 Å resolution structure of the pentameric procapsid protease of bacteriophage T4, gene product (gp)21. The structure corresponds to the enzyme's pre-active state in which its N-terminal region blocks the catalytic center, demonstrating that the activation mechanism involves self-cleavage of nine N-terminal residues. We describe similarities and differences between T4 gp21 and related herpesvirus proteases. We found that gp21 and the herpesvirus proteases have similarity with proteins forming the tubes of phage tails and bacterial type VI secretion systems, suggesting their common evolutionary origin.
Organizational Affiliation:
Department of Biological Sciences, Hockmeyer Hall of Structural Biology, Purdue University, 240 South Martin Jischke Drive, West Lafayette, IN 47907, USA. Electronic address: afokine@purdue.edu.