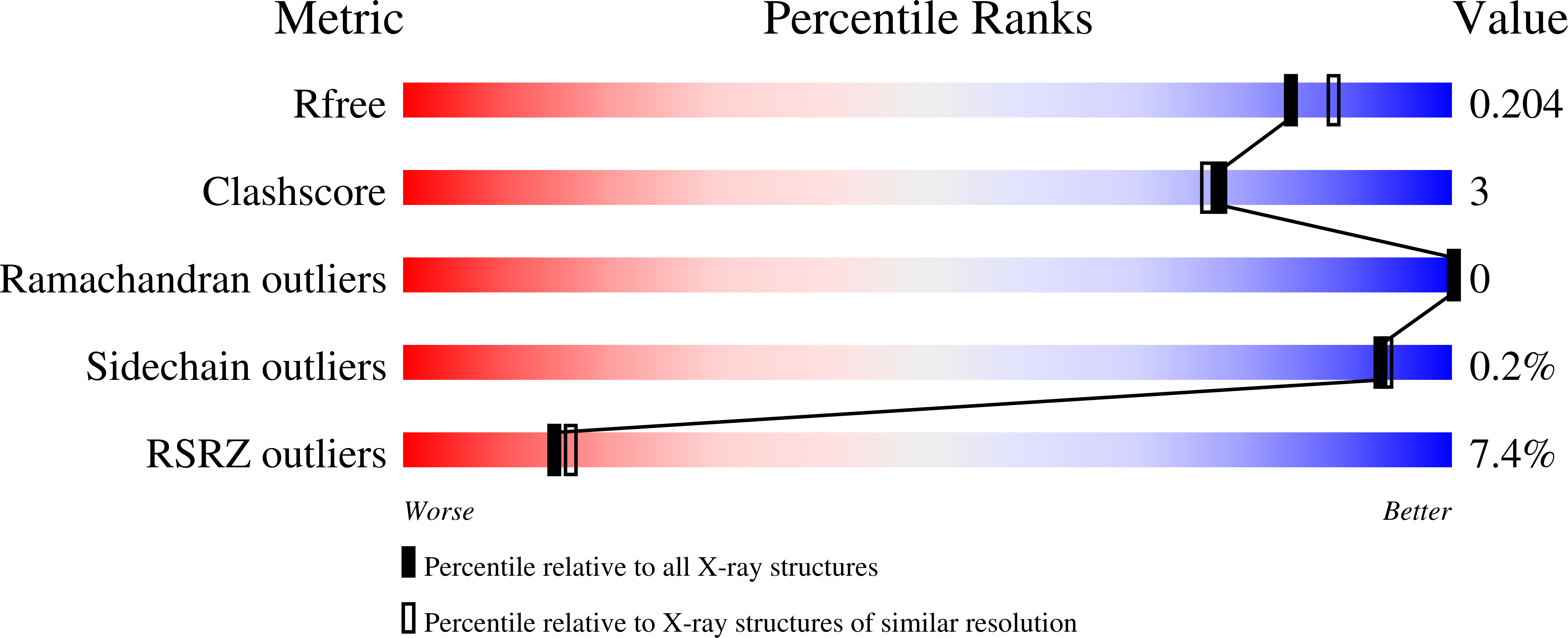
A New Member of the Thioredoxin Reductase Family from Early Oxygenic Photosynthetic Organisms.
Buey, R.M., Galindo-Trigo, S., Lopez-Maury, L., Velazquez-Campoy, A., Revuelta, J.L., Florencio, F.J., de Pereda, J.M., Schurmann, P., Buchanan, B.B., Balsera, M.(2017) Mol Plant 10: 212-215
- PubMed: 27418374
- DOI: https://doi.org/10.1016/j.molp.2016.06.019
- Primary Citation of Related Structures:
5J60
Organizational Affiliation:
Department Microbiología y Genética, Universidad de Salamanca, Salamanca 37007, Spain.