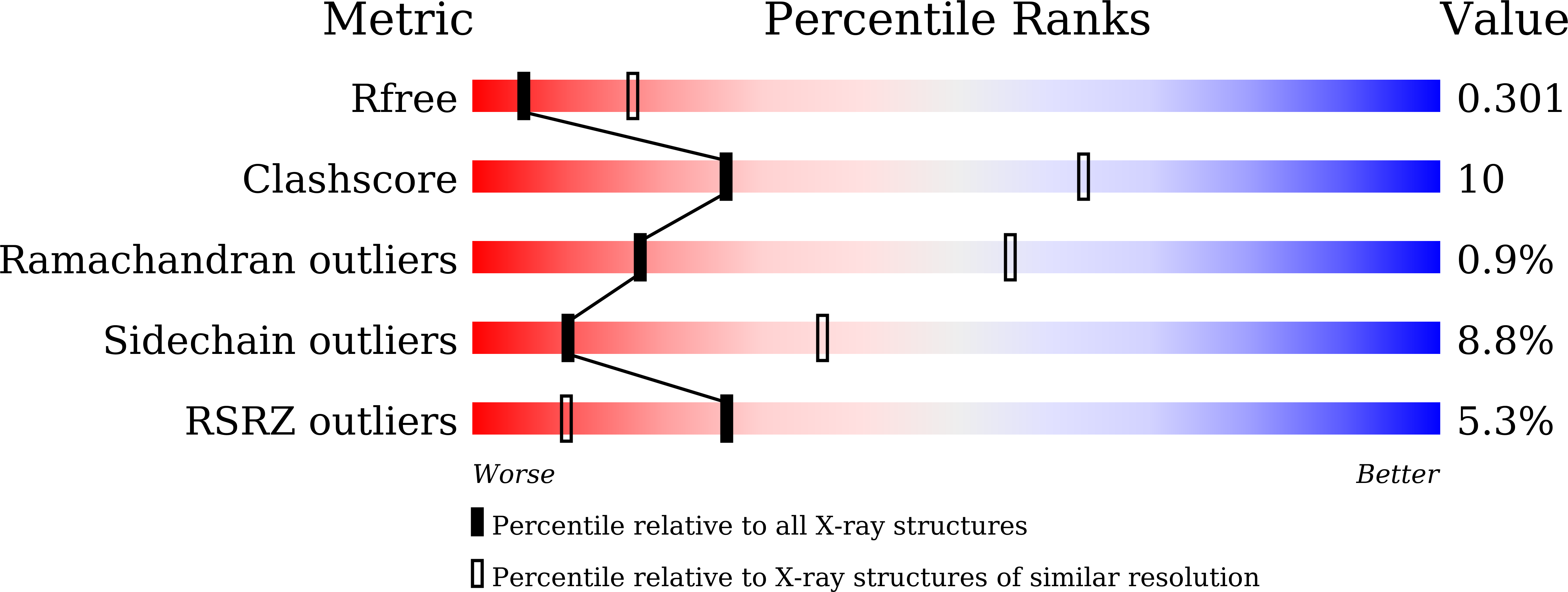
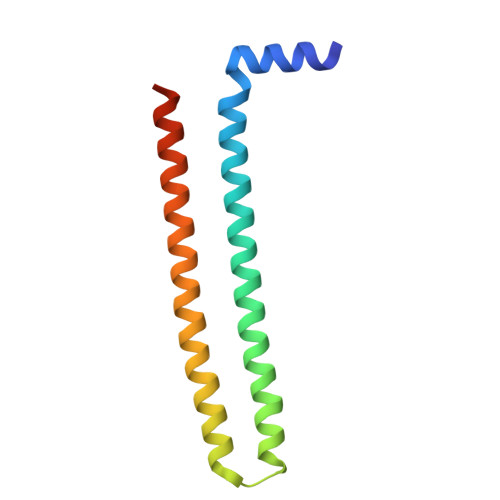
Structural Insights on PHA Binding Protein PhaP from Aeromonas hydrophila
Zhao, H., Wei, H., Liu, X., Yao, Z., Xu, M., Wei, D., Wang, J., Wang, X., Chen, G.Q.(2016) Sci Rep 6: 39424-39424
- PubMed: 28009010
- DOI: https://doi.org/10.1038/srep39424
- Primary Citation of Related Structures:
5IP0 - PubMed Abstract:
Phasins or PhaPs are a group of amphiphilic proteins that are found attached to the surface of microbial polyhydroxyalkanoate (PHA) granules. They have both structural and regulatory functions and can affect intracellular PHA accumulation and mediate protein folding. The molecular basis for the diverse functions of the PhaPs has not been fully understood due to the lack of the structural knowledge. Here we report the structural and biochemical studies of the PhaP cloned from Aeromonas hydrophila (PhaP Ah ), which is utilized in protein and tissue engineering. The crystal structure of PhaP Ah was revealed to be a tetramer with 8 α-helices adopting a coiled-coil structure. Each monomer has a hydrophobic and a hydrophilic surface, rendering the surfactant properties of the PhaP Ah monomer. Based on the crystal structure, we predicted three key amino acid residues and obtained mutants with enhanced stability and improved emulsification properties. The first PhaP crystal structure, as reported in this study, is an important step towards a mechanistic understanding of how PHA is formed in vivo and why PhaP has such unique surfactant properties. At the same time, it will facilitate the study of other PhaP members that may have significant biotechnological potential as bio-surfactants and amphipathic coatings.
Organizational Affiliation:
Center for Synthetic and Systems Biology, School of Life Science, Tsinghua-Peking Center for Life Sciences, Tsinghua University, Beijing 100084, China.