Crystal structure of the C-terminal Domain of Coenzyme A biosynthesis bifunctional protein CoaBC
Nocek, B., Zhou, M., Grimshaw, S., Anderson, W.F., Joachimiak, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Phosphopantothenate--cysteine ligase | 220 | Bacillus anthracis | Mutation(s): 0 Gene Names: coaBC, GBAA_4007, AB163_23415, AB164_18935, AB165_13850, AB166_05815, AB167_23710, AB168_27015, AB169_03120, AB170_11765... EC: 6.3.2.5 | 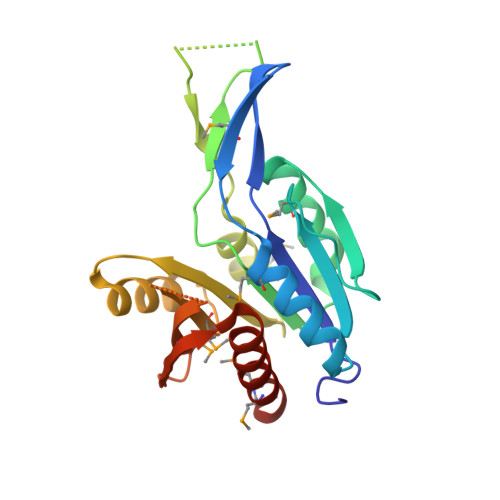 | |
UniProt | |||||
Find proteins for A0A6L8Q0G8 (Bacillus anthracis) Explore A0A6L8Q0G8 Go to UniProtKB: A0A6L8Q0G8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A6L8Q0G8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 133.173 | α = 90 |
| b = 133.173 | β = 90 |
| c = 55.765 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| HKL-3000 | phasing |