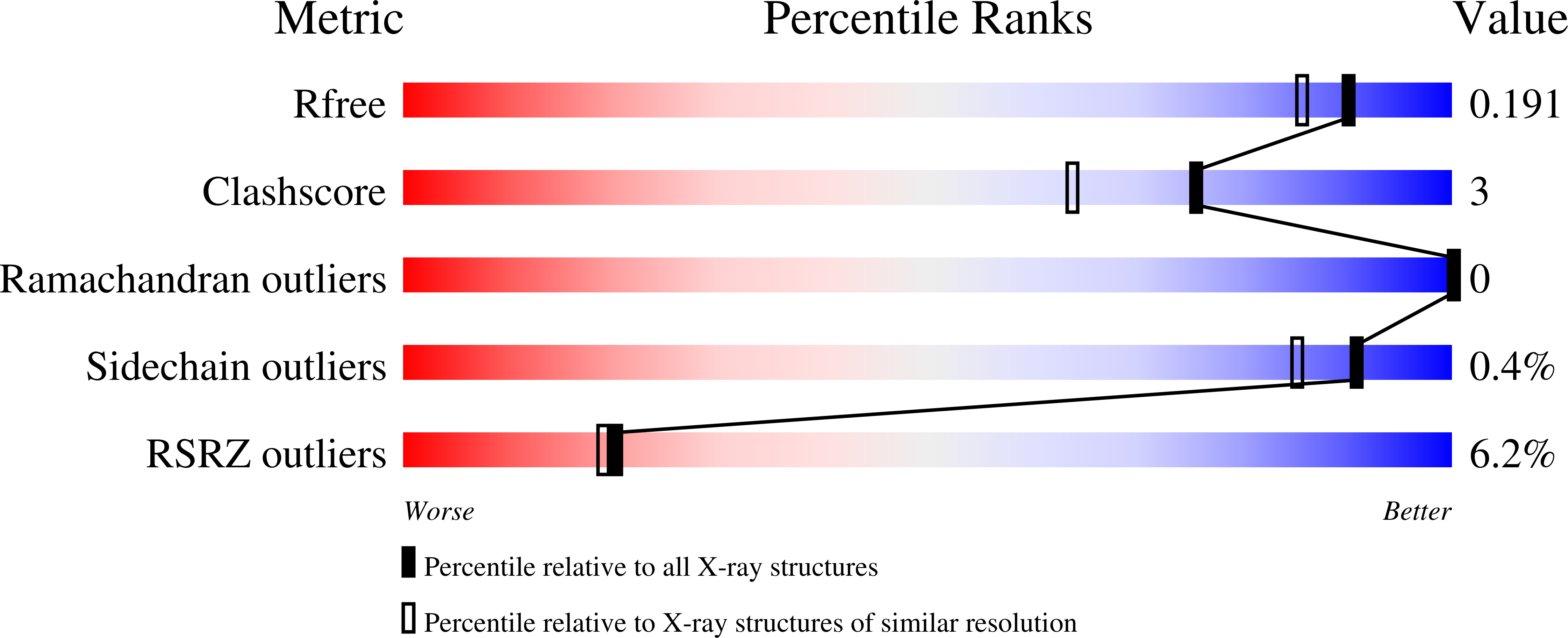
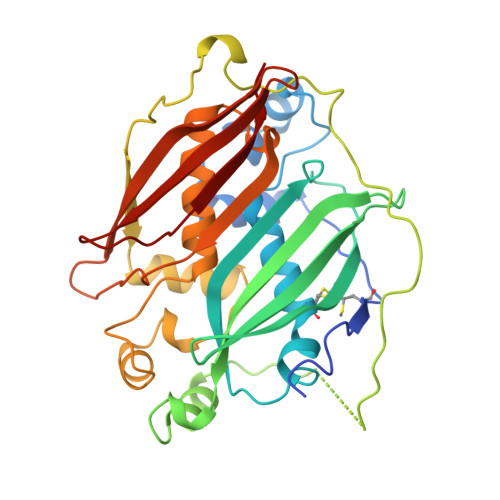
Deletion of a dehydratase important for intracellular growth and cording renders rough Mycobacterium abscessus avirulent.
Halloum, I., Carrere-Kremer, S., Blaise, M., Viljoen, A., Bernut, A., Le Moigne, V., Vilcheze, C., Guerardel, Y., Lutfalla, G., Herrmann, J.L., Jacobs, W.R., Kremer, L.(2016) Proc Natl Acad Sci U S A 113: E4228-E4237
- PubMed: 27385830
- DOI: https://doi.org/10.1073/pnas.1605477113
- Primary Citation of Related Structures:
5I7N - PubMed Abstract:
Mycobacterium abscessus (Mabs) is a rapidly growing Mycobacterium and an emerging pathogen in humans. Transitioning from a smooth (S) high-glycopeptidolipid (GPL) producer to a rough (R) low-GPL producer is associated with increased virulence in zebrafish, which involves the formation of massive serpentine cords, abscesses, and rapid larval death. Generating a cord-deficient Mabs mutant would allow us to address the contribution of cording in the physiopathological signs of the R variant. Herein, a deletion mutant of MAB_4780, encoding a dehydratase, distinct from the β-hydroxyacyl-ACP dehydratase HadABC complex, was constructed in the R morphotype. This mutant exhibited an alteration of the mycolic acid composition and a pronounced defect in cording. This correlated with an extremely attenuated phenotype not only in wild-type but also in immunocompromised zebrafish embryos lacking either macrophages or neutrophils. The abolition of granuloma formation in embryos infected with the dehydratase mutant was associated with a failure to replicate in macrophages, presumably due to limited inhibition of the phagolysosomal fusion. Overall, these results indicate that MAB_4780 is required for Mabs to successfully establish acute and lethal infections. Therefore, targeting MAB_4780 may represent an attractive antivirulence strategy to control Mabs infections, refractory to most standard chemotherapeutic interventions. The combination of a dehydratase assay with a high-resolution crystal structure of MAB_4780 opens the way to identify such specific inhibitors.
Organizational Affiliation:
Centre d'études d'agents Pathogènes et Biotechnologies pour la Santé (CPBS), CNRS Formation de Recherche en Evolution 3689, 34293 Montpellier, France;