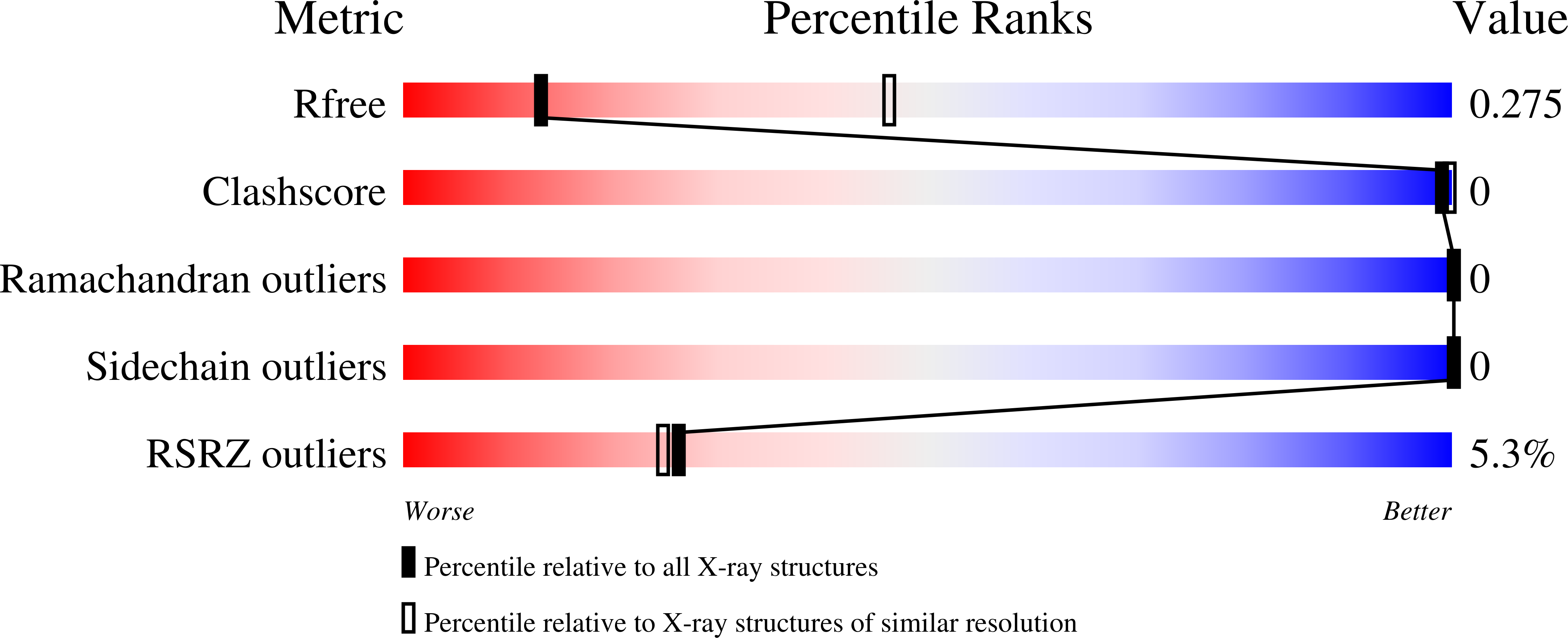
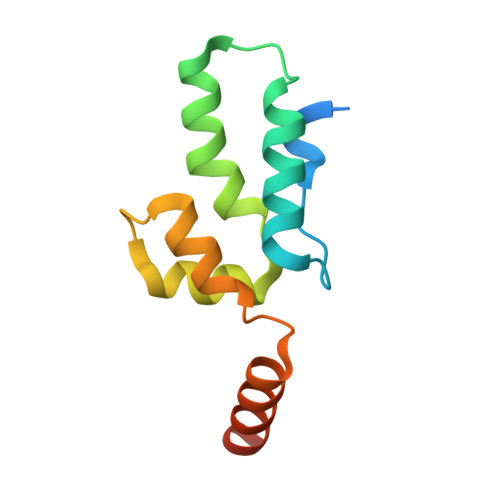
Crystal structure of an HIV assembly and maturation switch.
Wagner, J.M., Zadrozny, K.K., Chrustowicz, J., Purdy, M.D., Yeager, M., Ganser-Pornillos, B.K., Pornillos, O.(2016) Elife 5
- PubMed: 27416583
- DOI: https://doi.org/10.7554/eLife.17063
- Primary Citation of Related Structures:
5I4T - PubMed Abstract:
Virus assembly and maturation proceed through the programmed operation of molecular switches, which trigger both local and global structural rearrangements to produce infectious particles. HIV-1 contains an assembly and maturation switch that spans the C-terminal domain (CTD) of the capsid (CA) region and the first spacer peptide (SP1) of the precursor structural protein, Gag. The crystal structure of the CTD-SP1 Gag fragment is a goblet-shaped hexamer in which the cup comprises the CTD and an ensuing type II β-turn, and the stem comprises a 6-helix bundle. The β-turn is critical for immature virus assembly and the 6-helix bundle regulates proteolysis during maturation. This bipartite character explains why the SP1 spacer is a critical element of HIV-1 Gag but is not a universal property of retroviruses. Our results also indicate that HIV-1 maturation inhibitors suppress unfolding of the CA-SP1 junction and thereby delay access of the viral protease to its substrate.
Organizational Affiliation:
Department of Molecular Physiology and Biological Physics, University of Virginia, Charlottesville, United States.