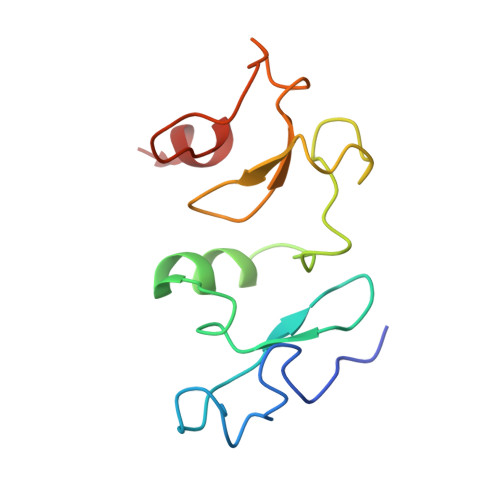
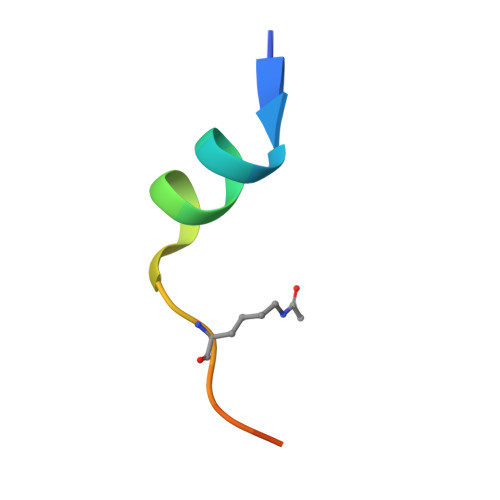
Crystal structure of DPF3b in complex with an acetylated histone peptide.
Li, W., Zhao, A., Tempel, W., Loppnau, P., Liu, Y.(2016) J Struct Biol 195: 365-372
- PubMed: 27402533
- DOI: https://doi.org/10.1016/j.jsb.2016.07.001
- Primary Citation of Related Structures:
5I3L - PubMed Abstract:
Histone acetylation plays an important role in chromatin dynamics and is associated with active gene transcription. This modification is written by acetyltransferases, erased by histone deacetylases and read out by bromodomain containing proteins, and others such as tandem PHD fingers of DPF3b. Here we report the high resolution crystal structure of the tandem PHD fingers of DPF3b in complex with an H3K14ac peptide. In the complex structure, the histone peptide adopts an α-helical conformation, unlike previously observed by NMR, but similar to a previously reported MOZ-H3K14ac complex structure. Our crystal structure adds to existing evidence that points to the α-helix as a natural conformation of histone tails as they interact with histone-associated proteins.
Organizational Affiliation:
Key Laboratory of Pesticide & Chemical Biology, College of Chemistry, Central China Normal University, Wuhan 430079, PR China; Structural Genomics Consortium, University of Toronto, 101 College Street, Toronto, Ontario M5G 1L7, Canada.