Structure of chromatin remodeler Swi2/Snf2 in the resting state
Xia, X., Liu, X., Li, T., Fang, X., Chen, Z.C.(2016) Nat Struct Mol Biol 23: 722-729
- PubMed: 27399259
- DOI: https://doi.org/10.1038/nsmb.3259
- Primary Citation of Related Structures:
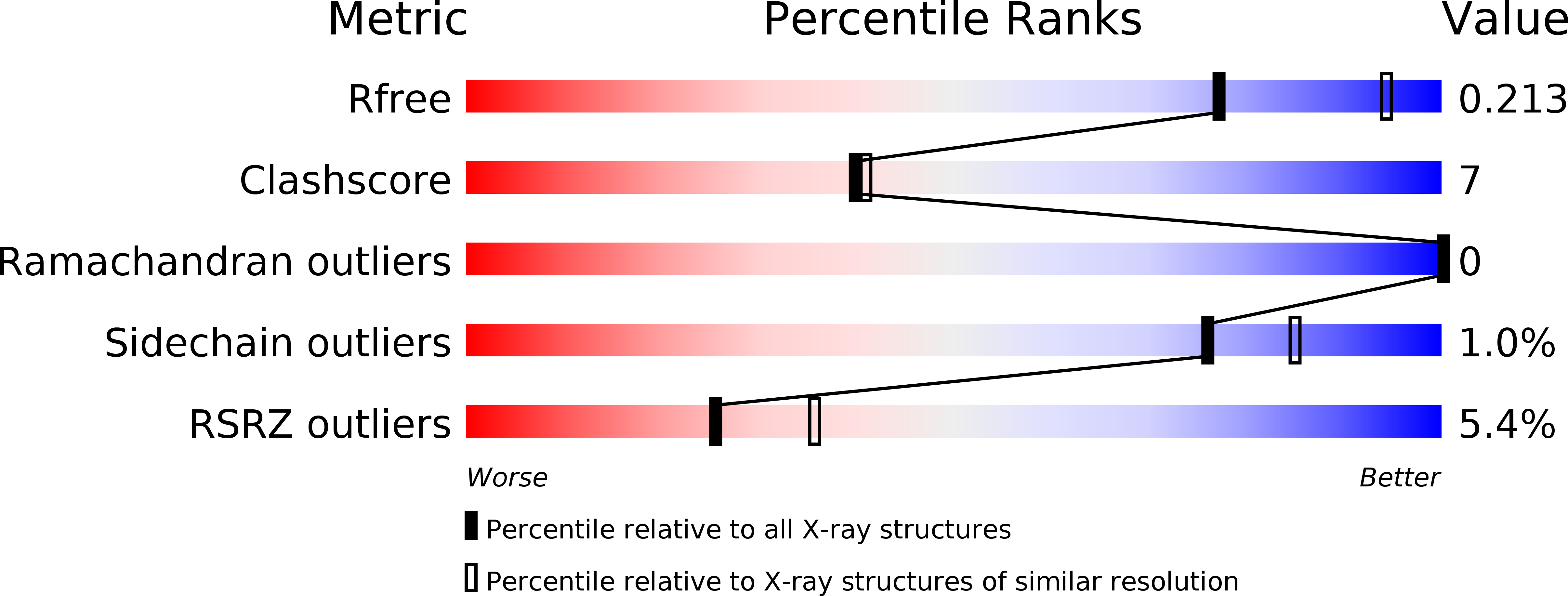
5HZR - PubMed Abstract:
SWI2/SNF2 family proteins regulate a myriad of nucleic acid transactions by sliding, removing and reconstructing nucleosomes in eukaryotic cells. They contain two RecA-like core domains, which couple ATP hydrolysis and DNA translocation to chromatin remodeling. Here we report the crystal structure of Snf2 from the yeast Myceliophthora thermophila. The data show the two RecA-like core domains of Snf2 stacking together and twisting their ATP-binding motifs away from each other, thus explaining the inactivity of the protein in the ground state. We identified several DNA-binding elements, which are fully exposed to solvent, thus suggesting that the protein is poised for its incoming substrate. The catalytic core of Snf2 showed a high chromatin-remodeling activity, which was suppressed by the N-terminal HSA domain. Our findings reveal that the catalytic core of Snf2 is a competent remodeling machine, which rests in an inactive conformation and requires a large conformational change upon activation.
Organizational Affiliation:
MOE Key Laboratory of Protein Science, Tsinghua University, Beijing, China.