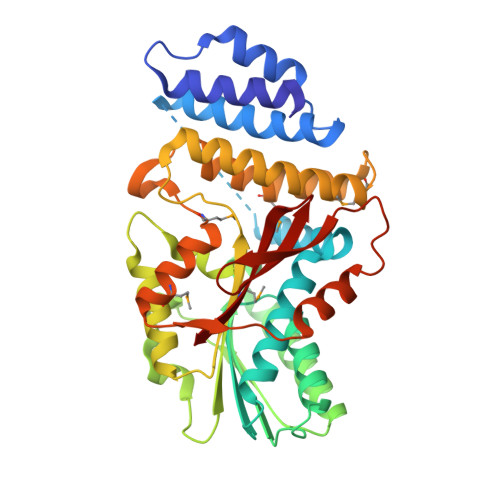
Crystal Structure of the GRAS Domain of SCARECROW-LIKE7 in Oryza sativa.
Li, S., Zhao, Y., Zhao, Z., Wu, X., Sun, L., Liu, Q., Wu, Y.(2016) Plant Cell 28: 1025-1034
- PubMed: 27081181
- DOI: https://doi.org/10.1105/tpc.16.00018
- Primary Citation of Related Structures:
5HYZ - PubMed Abstract:
GRAS proteins belong to a plant-specific protein family with many members and play essential roles in plant growth and development, functioning primarily in transcriptional regulation. Proteins in the family are minimally defined as containing the conserved GRAS domain. Here, we determined the structure of the GRAS domain of Os-SCL7 from rice (Oryza sativa) to 1.82 Å. The structure includes cap and core subdomains and elucidates the features of the conserved GRAS LRI, VHIID, LRII, PFYRE, and SAW motifs. The structure is a dimer, with a clear groove to accommodate double-stranded DNA. Docking a DNA segment into the groove to generate an Os-SCL7/DNA complex provides insight into the DNA binding mechanism of GRAS proteins. Furthermore, the in vitro DNA binding property of Os-SCL7 and model-defined recognition residues are assessed by electrophoretic mobility shift analysis and mutagenesis assays. These studies reveal the structure and preliminary DNA interaction mechanisms of GRAS proteins and open the door to in-depth investigation and understanding of the individual pathways in which they play important roles.
Organizational Affiliation:
State Key Laboratory of Structural Chemistry, Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences, Fuzhou 350002, China.