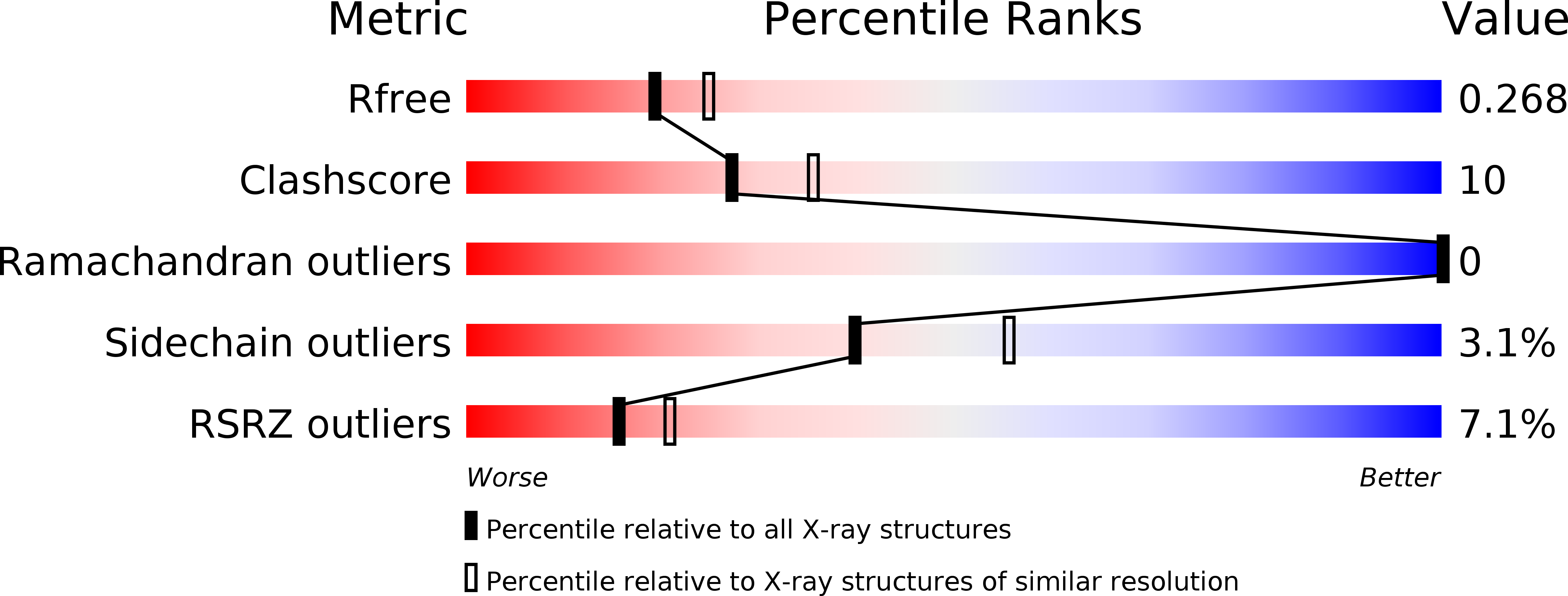
Solving the crystal structure of human calcium-free S100Z: the siege and conquer of one of the last S100 family strongholds.
Calderone, V., Fragai, M., Gallo, G., Luchinat, C.(2017) J Biol Inorg Chem 22: 519-526
- PubMed: 28074300
- DOI: https://doi.org/10.1007/s00775-017-1437-4
- Primary Citation of Related Structures:
5HYD - PubMed Abstract:
The X-ray structure of human apo-S100Z has been solved and compared with that of the zebrafish calcium-bound S100Z, which is the closest in sequence. Human apo-S100A12, which shows only 43% sequence identity to human S100Z, has been used as template model to solve the crystallographic phase problem. Although a significant buried surface area between the two physiological dimers is present in the asymmetric unit of human apo-S100Z, the protein does not form the superhelical arrangement in the crystal as observed for the zebrafish calcium-bound S100Z and human calcium-bound S100A4. These findings further demonstrate that calcium plays a fundamental role in triggering quaternary structure formation in several S100s. Solving the X-ray structure of human apo-S100Z by standard molecular replacement procedures turned out to be a challenge and required trying different models and different software tools among which only one was successful. The model that allowed structure solution was that with one of the lowest sequence identity with the target protein among the S100 family in the apo state. Based on the previously solved zebrafish holo-S100Z, a putative human holo-S100Z structure has been then calculated through homology modeling; the differences between the experimental human apo and calculated holo structure have been compared to those existing for other members of the family.
Organizational Affiliation:
CERM, University of Florence, via Luigi Sacconi, 6, 50019, Sesto Fiorentino, FI, Italy. calderone@cerm.unifi.it.