Fine Tuning of Chlorophyll Spectra by Protein-Induced Ring Deformation.
Bednarczyk, D., Dym, O., Prabahar, V., Peleg, Y., Pike, D.H., Noy, D.(2016) Angew Chem Int Ed Engl 55: 6901-6905
- PubMed: 27098554
- DOI: https://doi.org/10.1002/anie.201512001
- Primary Citation of Related Structures:
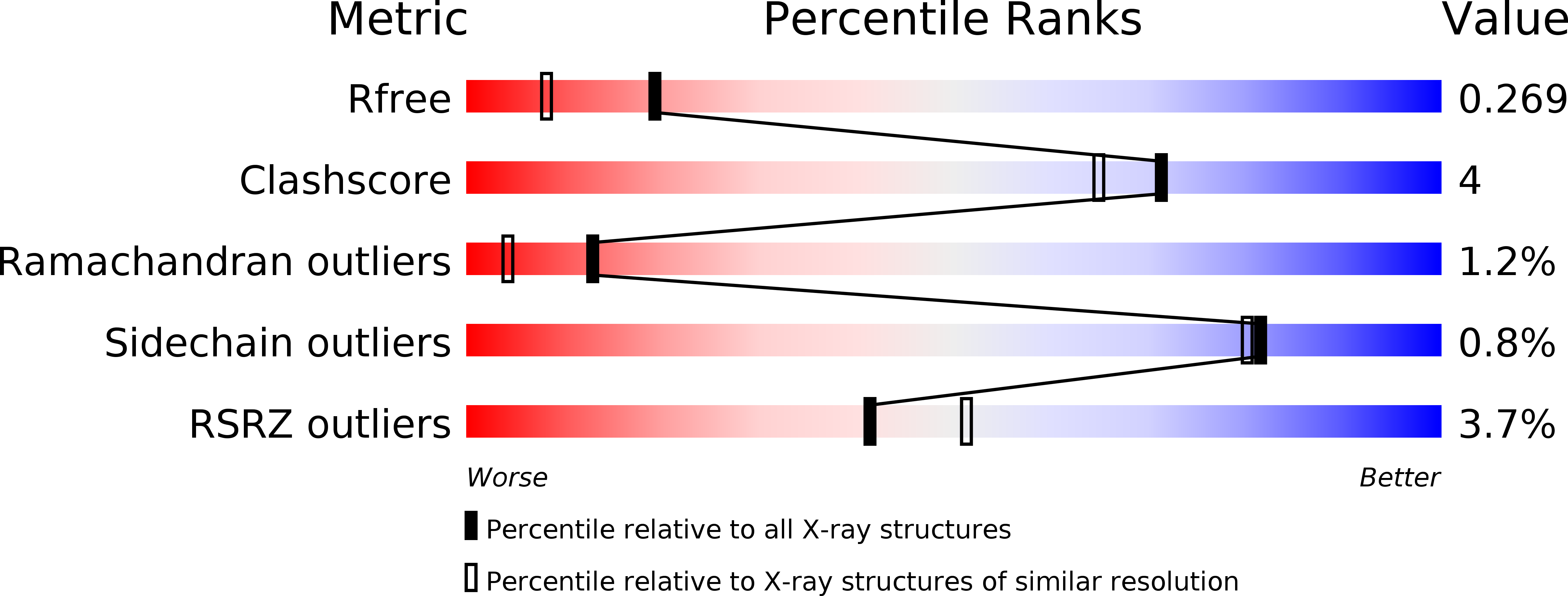
5HPZ - PubMed Abstract:
The ability to tune the light-absorption properties of chlorophylls by their protein environment is the key to the robustness and high efficiency of photosynthetic light-harvesting proteins. Unfortunately, the intricacy of the natural complexes makes it very difficult to identify and isolate specific protein-pigment interactions that underlie the spectral-tuning mechanisms. Herein we identify and demonstrate the tuning mechanism of chlorophyll spectra in type II water-soluble chlorophyll binding proteins from Brassicaceae (WSCPs). By comparing the molecular structures of two natural WSCPs we correlate a shift in the chlorophyll red absorption band with deformation of its tetrapyrrole macrocycle that is induced by changing the position of a nearby tryptophan residue. We show by a set of reciprocal point mutations that this change accounts for up to 2/3 of the observed spectral shift between the two natural variants.
Organizational Affiliation:
Department of Biological Chemistry, Weizmann Institute of Science, Rehovot, Israel.