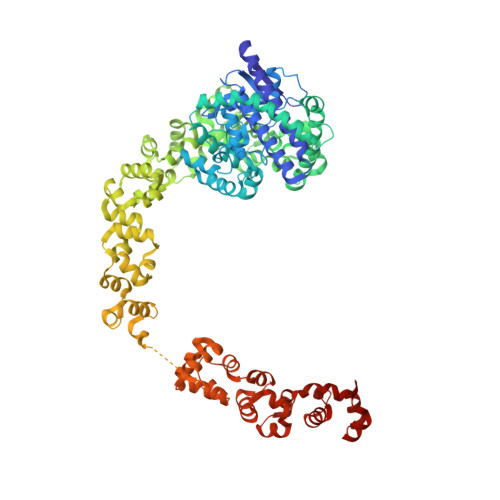
Methanopyrus kandleri topoisomerase V contains three distinct AP lyase active sites in addition to the topoisomerase active site.
Rajan, R., Osterman, A., Mondragon, A.(2016) Nucleic Acids Res 44: 3464-3474
- PubMed: 26908655
- DOI: https://doi.org/10.1093/nar/gkw122
- Primary Citation of Related Structures:
5HM5 - PubMed Abstract:
Topoisomerase V (Topo-V) is the only topoisomerase with both topoisomerase and DNA repair activities. The topoisomerase activity is conferred by a small alpha-helical domain, whereas the AP lyase activity is found in a region formed by 12 tandem helix-hairpin-helix ((HhH)2) domains. Although it was known that Topo-V has multiple repair sites, only one had been mapped. Here, we show that Topo-V has three AP lyase sites. The atomic structure and Small Angle X-ray Scattering studies of a 97 kDa fragment spanning the topoisomerase and 10 (HhH)2 domains reveal that the (HhH)2 domains extend away from the topoisomerase domain. A combination of biochemical and structural observations allow the mapping of the second repair site to the junction of the 9th and 10th (HhH)2 domains. The second site is structurally similar to the first one and to the sites found in other AP lyases. The 3rd AP lyase site is located in the 12th (HhH)2 domain. The results show that Topo-V is an unusual protein: it is the only known protein with more than one (HhH)2 domain, the only known topoisomerase with dual activities and is also unique by having three AP lyase repair sites in the same polypeptide.
Organizational Affiliation:
Department of Molecular Biosciences, Northwestern University, 2205 Tech Drive, Evanston, IL 60208, USA.