Structural Basis for the Unique Multivalent Readout of Unmodified H3 Tail by Arabidopsis ORC1b BAH-PHD Cassette
Li, S., Yang, Z., Du, X., Liu, R., Wilkinson, A.W., Gozani, O., Jacobsen, S.E., Patel, D.J., Du, J.(2016) Structure 24: 486-494
- PubMed: 26876097
- DOI: https://doi.org/10.1016/j.str.2016.01.004
- Primary Citation of Related Structures:
5HH7 - PubMed Abstract:
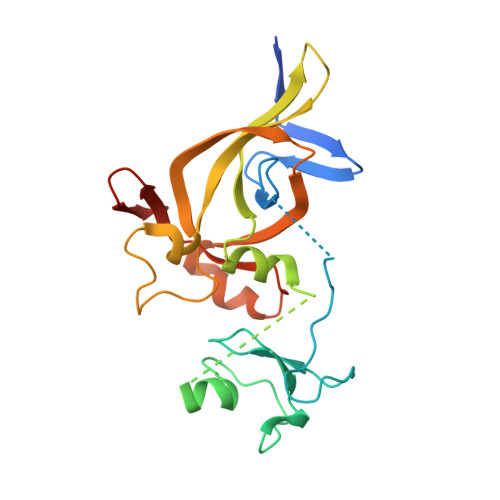
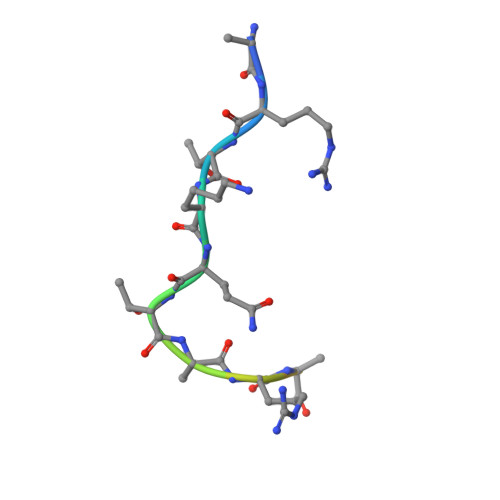
DNA replication initiation relies on the formation of the origin recognition complex (ORC). The plant ORC subunit 1 (ORC1) protein possesses a conserved N-terminal BAH domain with an embedded plant-specific PHD finger, whose function may be potentially regulated by an epigenetic mechanism. Here, we report structural and biochemical studies on the Arabidopsis thaliana ORC1b BAH-PHD cassette which specifically recognizes the unmodified H3 tail. The crystal structure of ORC1b BAH-PHD cassette in complex with an H3(1-15) peptide reveals a strict requirement for the unmodified state of R2, T3, and K4 on the H3 tail and a novel multivalent BAH and PHD readout mode for H3 peptide recognition. Such recognition may contribute to epigenetic regulation of the initiation of DNA replication.
Organizational Affiliation:
Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10065, USA.