NMR studies on the interactions between yeast Vta1 and Did2 during the multivesicular bodies sorting pathway
Shen, J., Yang, Z., Wang, J., Zhao, B., Lan, W., Wang, C., Zhang, X., Wild, C.J., Liu, M., Xu, Z., Cao, C.(2016) Sci Rep 6: 38710-38710
- PubMed: 27924850
- DOI: https://doi.org/10.1038/srep38710
- Primary Citation of Related Structures:
5H7P - PubMed Abstract:
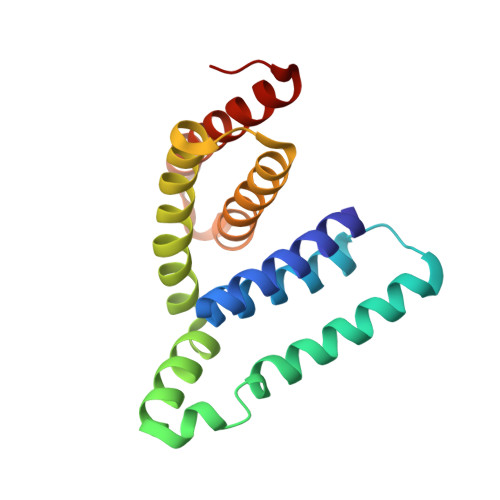
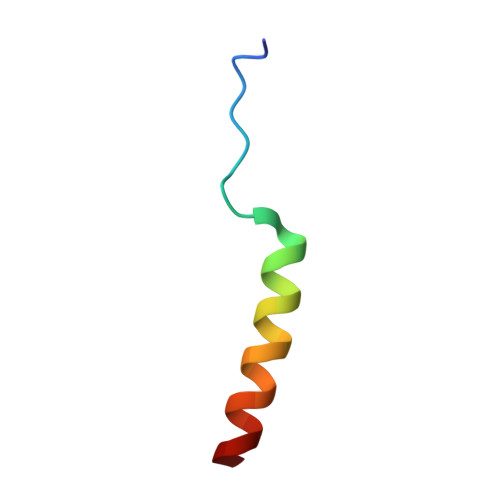
As an AAA-ATPase, Vps4 is important for function of multivesicular bodies (MVB) sorting pathway, which involves in cellular phenomena ranging from receptor down-regulation to viral budding to cytokinesis. The activity of Vps4 is stimulated by the interactions between Vta1 N-terminus (named as Vta1NTD) and Did2 fragment (176-204 aa) (termed as Did2 176-204 ) or Vps60 (128-186 aa) (termed as Vps60 128-186 ). The structural basis of how Vta1NTD binds to Did2 176-204 is still unclear. To address this, in this report, the structure of Did2 176-204 in complex with Vta1NTD was determined by NMR techniques, demonstrating that Did2 176-204 interacts with Vta1NTD through its helix α6' extending over the 2 nd and the 3 rd α-helices of Vta1NTD microtubule interacting and transport 1 (MIT1) domain. The residues within Did2 176-204 helix α6' in the interface make up of an amino acid sequence as E 192 'xxL 195 'xxR 198 'L 199 'xxL 202 'R 203 ', identical to type 1 MIT-interacting motif (MIM1) (D/E)xxLxxRLxxL(K/R) of CHMP1A 180-196 observed in Vps4-CHMP1A complex structure, indicating that Did2 binds to Vta1NTD through canonical MIM1 interactions. Moreover, the Did2 binding does not result in Vta1NTD significant conformational changes, revealing that Did2, similar to Vps60, enhances Vta1 stimulation of Vps4 ATPase activity in an indirect manner.
Organizational Affiliation:
State Key Laboratory of Bioorganic and Natural Products Chemistry and Collaborative Innovation Center of Chemistry for Life Sciences, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, 345 Lingling Road, Shanghai, 200032, China.