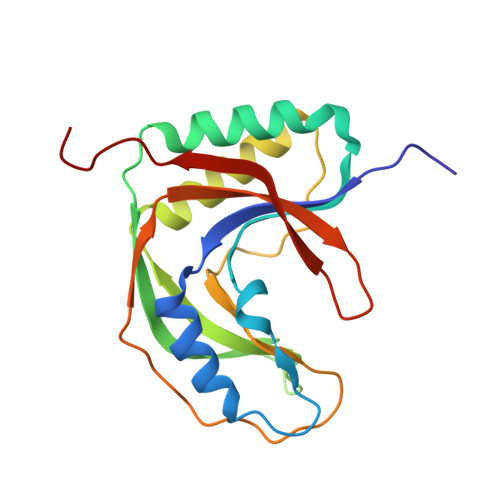
Crystal structure of the RNA 2',3'-cyclic phosphodiesterase from Deinococcus radiodurans
Han, W., Cheng, J., Zhou, C., Hua, Y., Zhao, Y.(2017) Acta Crystallogr F Struct Biol Commun 73: 276-280
- PubMed: 28471359
- DOI: https://doi.org/10.1107/S2053230X17004964
- Primary Citation of Related Structures:
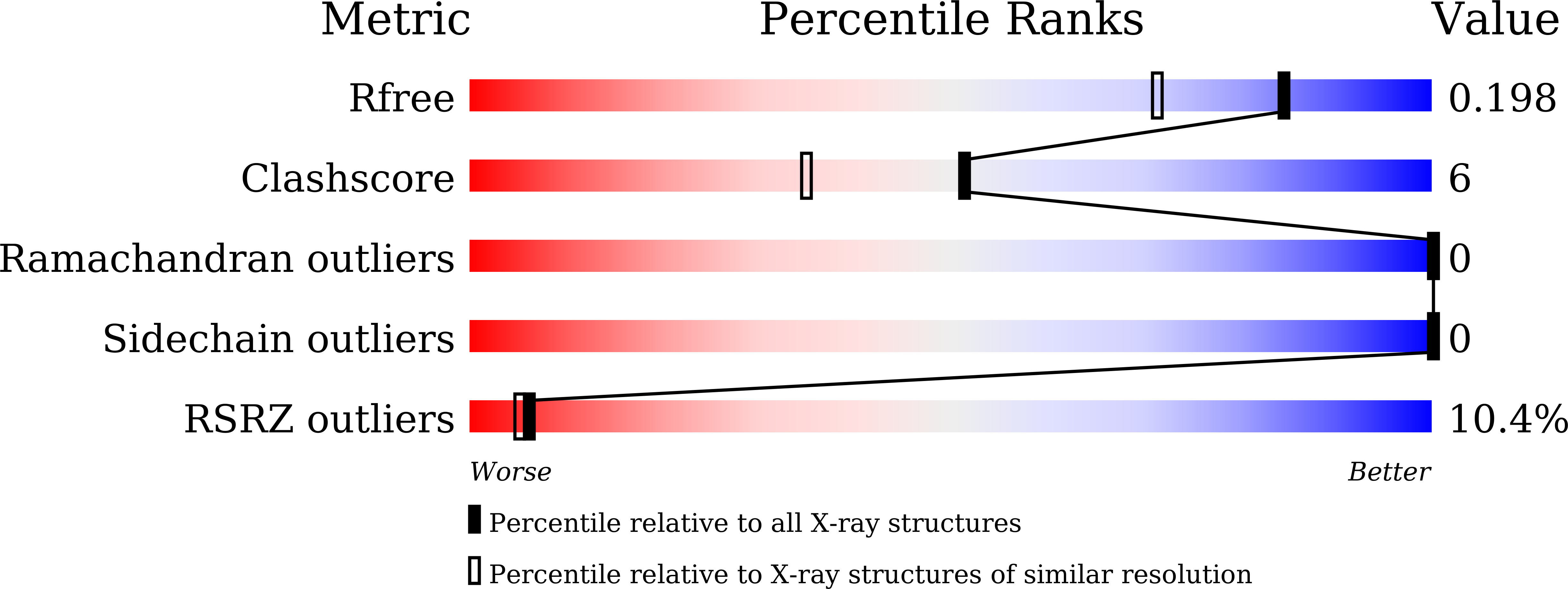
5H7E - PubMed Abstract:
2',3'-Cyclic phosphodiesterase (CPDase) homologues have been found in all domains of life and are involved in diverse RNA and nucleotide metabolisms. The CPDase from Deinococcus radiodurans was crystallized and the crystals diffracted to 1.6 Å resolution, which is the highest resolution currently known for a CPDase structure. Structural comparisons revealed that the enzyme is in an open conformation in the absence of substrate. Nevertheless, the active site is well formed, and the representative motifs interact with sulfate ion, which suggests a conserved catalytic mechanism.
Organizational Affiliation:
Key Laboratory of Chinese Ministry of Agriculture for Nuclear-Agricultural Sciences, Institute of Nuclear-Agricultural Sciences, Zhejiang University, People's Republic of China.