Structural basis of the substrate recognition of hydrazidase isolated from Microbacterium sp. strain HM58-2, which catalyzes acylhydrazide compounds as its sole carbon source
Akiyama, T., Ishii, M., Takuwa, A., Oinuma, K.I., Sasaki, Y., Takaya, N., Yajima, S.(2017) Biochem Biophys Res Commun 482: 1007-1012
- PubMed: 27908731
- DOI: https://doi.org/10.1016/j.bbrc.2016.11.148
- Primary Citation of Related Structures:
5H6S, 5H6T - PubMed Abstract:
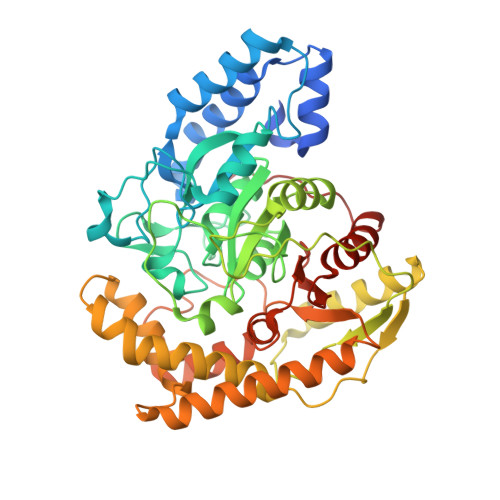
Hydrazidase was an enzyme that remained unidentified for a half century. However, recently, it was purified, and its encoding gene was cloned. Microbacterium sp. strain HM58-2 grows with acylhydrazides as its sole carbon source; it produces hydrazidase and degrades acylhydrazides to acetate and hydrazides. The bacterial hydrazidase belongs to the amidase signature enzyme family and contains a Ser-cisSer-Lys catalytic motif. The condensation of hydrazine and carbonic acid produces various hydrazides, some of which are raw materials for synthesizing pharmaceuticals and other useful chemicals. Although natural hydrazide compounds have been identified, the metabolic systems for hydrazides are not fully understood. Here, we report the crystal structure of hydrazidase from Microbacterium sp. strain HM58-2. The active site was revealed to consist of a Ser-cisSer-Lys catalytic triad, in which Ser179 forms a covalent bond with a carbonyl carbon of the substrate. 4-Hydroxybenzoic acid hydrazide bound to the S179A mutant, showing an oxyanion hole composed of the three backbone amide groups. Furthermore, H336 in the non-conserved region in the amidase family may define the substrate specificity, which was confirmed by mutation analysis. A wild-type apoenzyme structure revealed an unidentified molecule covalently bound to S179, representing a tetrahedral intermediate.
Organizational Affiliation:
Department of Bioscience, Tokyo University of Agriculture, Setagaya-ku, Tokyo 156-8502, Japan.