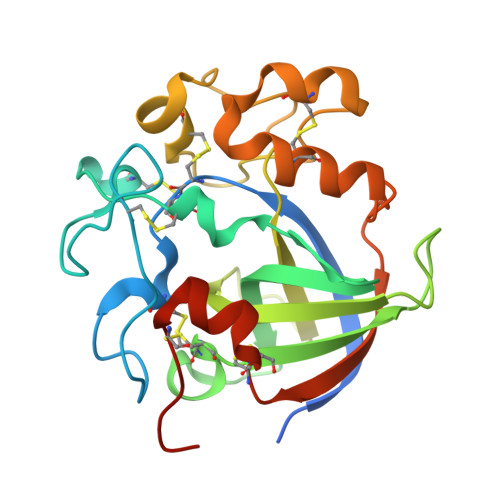
Genetic and Structural Characterization of a Thermo-Tolerant, Cold-Active, and Acidic Endo-beta-1,4-glucanase from Antarctic Springtail, Cryptopygus antarcticus.
Song, J.M., Hong, S.K., An, Y.J., Kang, M.H., Hong, K.H., Lee, Y.H., Cha, S.S.(2017) J Agric Food Chem 65: 1630-1640
- PubMed: 28156112
- DOI: https://doi.org/10.1021/acs.jafc.6b05037
- Primary Citation of Related Structures:
5H4U - PubMed Abstract:
The CaCel gene from Antarctic springtail Cryptopygus antarcticus codes for a cellulase belonging to the glycosyl hydrolase family 45 (GHF45). Phylogenetic, biochemical, and structural analyses revealed that the CaCel gene product (CaCel) is closely related to fungal GHF45 endo-β-1,4-glucanases. The organization of five introns within the open reading frame of the CaCel gene indicates its endogenous origin in the genome of the species, which suggests the horizontal transfer of the gene from fungi to the springtail. CaCel exhibited optimal activity at pH 3.5, retained 80% of its activity at 0-10 °C, and maintained a half-life of 4 h at 70 °C. Based on the structural comparison between CaCel and a fungal homologue, we deduced the structural basis for the unusual characteristics of CaCel. Under acidic conditions at 50 °C, CaCel was effective to digest the green algae (Ulva pertusa), suggesting that it could be exploited for biofuel production from seaweeds.
Organizational Affiliation:
Korea Institute of Ocean Science and Technology , 787 Haean-Ro, Sangnok-Gu, Ansan 426-744, Republic of Korea.