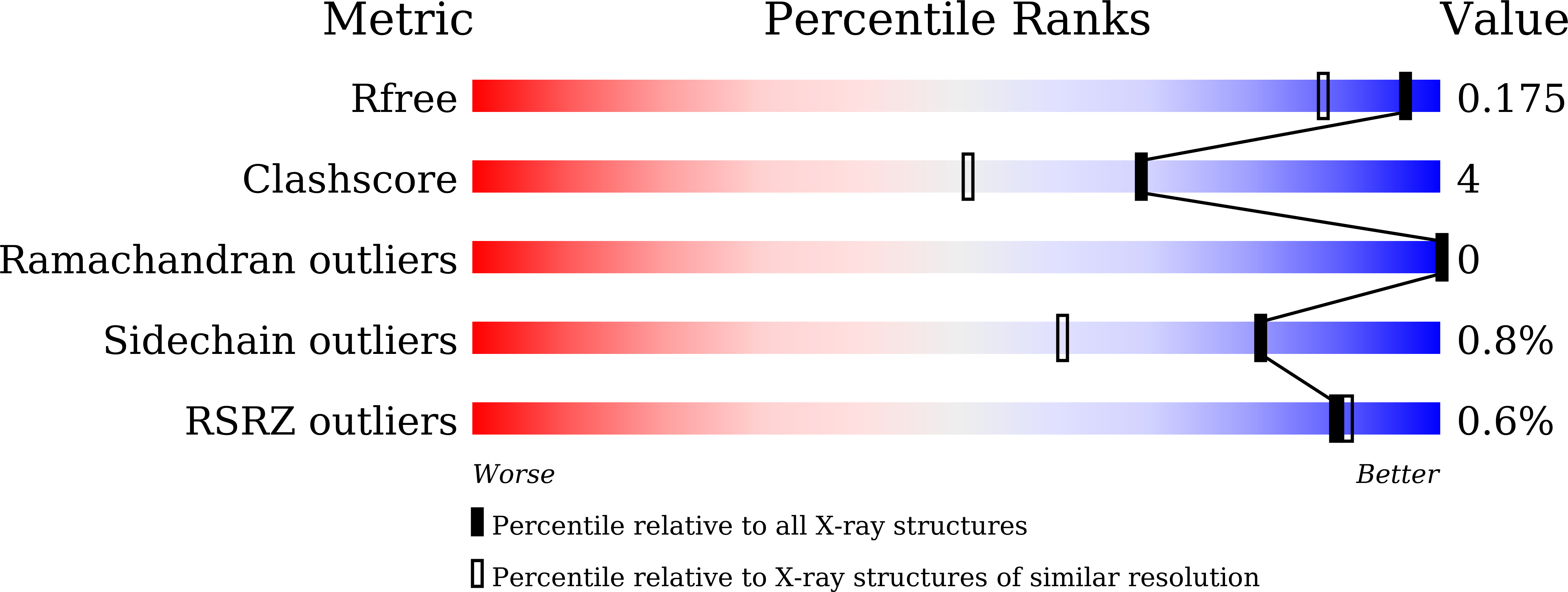
Structure-based engineering of a pectate lyase with improved specific activity for ramie degumming.
Zhou, Z., Liu, Y., Chang, Z., Wang, H., Leier, A., Marquez-Lago, T.T., Ma, Y., Li, J., Song, J.(2017) Appl Microbiol Biotechnol 101: 2919-2929
- PubMed: 28028551
- DOI: https://doi.org/10.1007/s00253-016-7994-6
- Primary Citation of Related Structures:
5GT5 - PubMed Abstract:
Biotechnological applications of microbial pectate lyases (Pels) in plant fiber processing are promising, eco-friendly substitutes for conventional chemical degumming processes. However, to potentiate the enzymes' use for industrial applications, resolving the molecular structure to elucidate catalytic mechanisms becomes necessary. In this manuscript, we report the high resolution (1.45 Å) crystal structure of pectate lyase (pelN) from Paenibacillus sp. 0602 in apo form. Through sequence alignment and structural superposition with other members of the polysaccharide lyase (PL) family 1 (PL1), we determined that pelN shares the characteristic right-handed β-helix and is structurally similar to other members of the PL1 family, while exhibiting key differences in terms of catalytic and substrate binding residues. Then, based on information from structure alignments with other PLs, we engineered a novel pelN. Our rational design yielded a pelN mutant with a temperature for enzymatic activity optimally shifted from 67.5 to 60 °C. Most importantly, this pelN mutant displayed both higher specific activity and ramie fiber degumming ability when compared with the wild-type enzyme. Altogether, our rational design method shows great potential for industrial applications. Moreover, we expect the reported high-resolution crystal structure to provide a solid foundation for future rational, structure-based engineering of genetically enhanced pelNs.
Organizational Affiliation:
National Engineering Laboratory for Industrial Enzymes and Key Laboratory of Systems Microbial Biotechnology, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin, 300308, China.