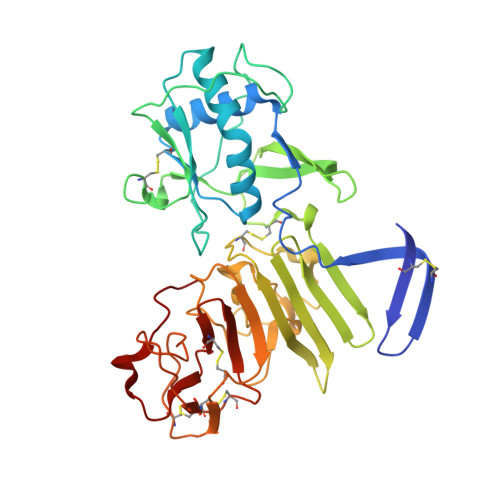
Contribution of intertwined loop to membrane association revealed by Zika virus full-length NS1 structure
Xu, X., Song, H., Qi, J., Liu, Y., Wang, H., Su, C., Shi, Y., Gao, G.F.(2016) EMBO J 35: 2170-2178
- PubMed: 27578809
- DOI: https://doi.org/10.15252/embj.201695290
- Primary Citation of Related Structures:
5GS6 - PubMed Abstract:
The association of Zika virus (ZIKV) infections with microcephaly and neurological diseases has highlighted an emerging public health concern. Here, we report the crystal structure of the full-length ZIKV nonstructural protein 1 (NS1), a major host-interaction molecule that functions in flaviviral replication, pathogenesis, and immune evasion. Of note, a long intertwined loop is observed in the wing domain of ZIKV NS1, and forms a hydrophobic "spike", which can contribute to cellular membrane association. For different flaviviruses, the amino acid sequences of the "spike" are variable but their common characteristic is either hydrophobic or positively charged, which is a beneficial feature for membrane binding. Comparative studies with West Nile and Dengue virus NS1 structures reveal conserved features, but diversified electrostatic characteristics on both inner and outer faces. Our results suggest different mechanisms of flavivirus pathogenesis and should be considered during the development of diagnostic tools.
Organizational Affiliation:
School of Life Sciences, University of Science and Technology of China, Hefei, China.