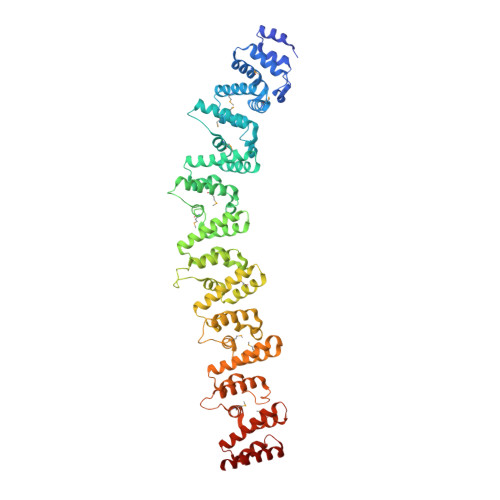
Structural analysis ofPhytophthorasuppressor of RNA silencing 2 (PSR2) reveals a conserved modular fold contributing to virulence.
He, J.Q., Ye, W.W., Choi, D.S., Wu, B.X., Zhai, Y., Guo, B.D., Duan, S.Y., Wang, Y.C., Gan, J.H., Ma, W.B., Ma, J.B.(2019) Proc Natl Acad Sci U S A
- PubMed: 30926664
- DOI: https://doi.org/10.1073/pnas.1819481116
- Primary Citation of Related Structures:
5GNC - PubMed Abstract:
Phytophthora are eukaryotic pathogens that cause enormous losses in agriculture and forestry. Each Phytophthora species encodes hundreds of effector proteins that collectively have essential roles in manipulating host cellular processes and facilitating disease development. Here we report the crystal structure of the effector Phytophthora suppressor of RNA silencing 2 (PSR2). PSR2 produced by the soybean pathogen Phytophthora sojae ( Ps PSR2) consists of seven tandem repeat units, including one W-Y motif and six L-W-Y motifs. Each L-W-Y motif forms a highly conserved fold consisting of five α-helices. Adjacent units are connected through stable, directional linkages between an internal loop at the C terminus of one unit and a hydrophobic pocket at the N terminus of the following unit. This unique concatenation results in an overall stick-like structure of Ps PSR2. Genome-wide analyses reveal 293 effectors from five Phytophthora species that have the Ps PSR2-like arrangement, that is, containing a W-Y motif as the "start" unit, various numbers of L-W-Y motifs as the "middle" units, and a degenerate L-W-Y as the "end" unit. Residues involved in the interunit interactions show significant conservation, suggesting that these effectors also use the conserved concatenation mechanism. Furthermore, functional analysis demonstrates differential contributions of individual units to the virulence activity of Ps PSR2. These findings suggest that the L-W-Y fold is a basic structural and functional module that may serve as a "building block" to accelerate effector evolution in Phytophthora .
Organizational Affiliation:
State Key Laboratory of Genetic Engineering, Ministry of Education Key Laboratory of Biodiversity Sciences and Ecological Engineering, Institute of Plant Biology, School of Life Sciences, Fudan University, 200438 Shanghai, China.