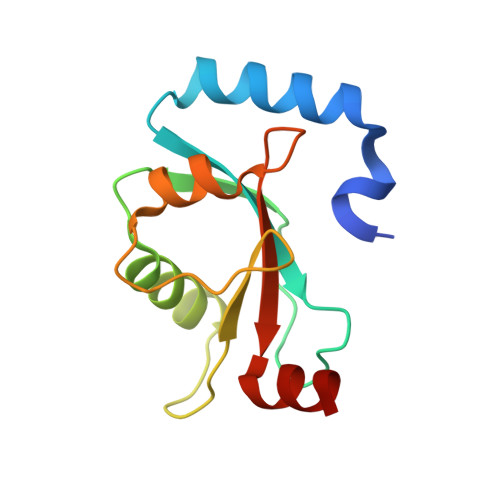
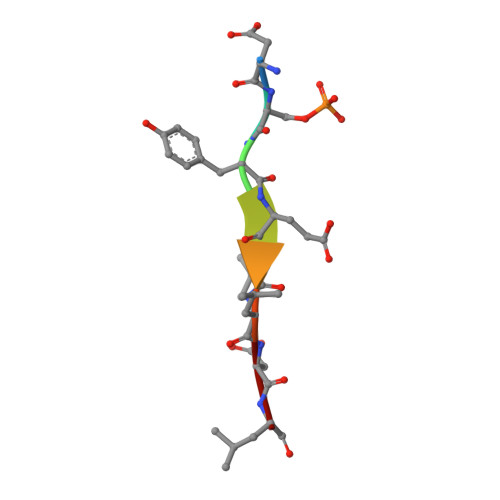
Structural insights into the recognition of phosphorylated FUNDC1 by LC3B in mitophagy
Lv, M., Wang, C., Li, F., Peng, J., Wen, B., Gong, Q., Shi, Y., Tang, Y.(2017) Protein Cell 8: 25-38
- PubMed: 27757847
- DOI: https://doi.org/10.1007/s13238-016-0328-8
- Primary Citation of Related Structures:
5GMV - PubMed Abstract:
Mitophagy is an essential intracellular process that eliminates dysfunctional mitochondria and maintains cellular homeostasis. Mitophagy is regulated by the post-translational modification of mitophagy receptors. Fun14 domain-containing protein 1 (FUNDC1) was reported to be a new receptor for hypoxia-induced mitophagy in mammalian cells and interact with microtubule-associated protein light chain 3 beta (LC3B) through its LC3 interaction region (LIR). Moreover, the phosphorylation modification of FUNDC1 affects its binding affinity for LC3B and regulates selective mitophagy. However, the structural basis of this regulation mechanism remains unclear. Here, we present the crystal structure of LC3B in complex with a FUNDC1 LIR peptide phosphorylated at Ser17 (pS 17 ), demonstrating the key residues of LC3B for the specific recognition of the phosphorylated or dephosphorylated FUNDC1. Intriguingly, the side chain of LC3B Lys49 shifts remarkably and forms a hydrogen bond and electrostatic interaction with the phosphate group of FUNDC1 pS 17 . Alternatively, phosphorylated Tyr18 (pY 18 ) and Ser13 (pS 13 ) in FUNDC1 significantly obstruct their interaction with the hydrophobic pocket and Arg10 of LC3B, respectively. Structural observations are further validated by mutation and isothermal titration calorimetry (ITC) assays. Therefore, our structural and biochemical results reveal a working model for the specific recognition of FUNDC1 by LC3B and imply that the reversible phosphorylation modification of mitophagy receptors may be a switch for selective mitophagy.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, 230027, China.