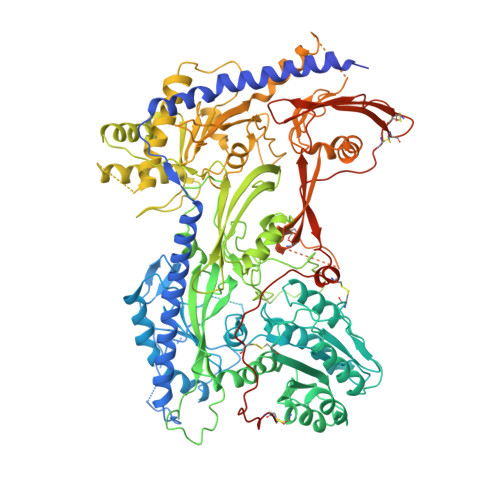
Structure of the voltage-gated calcium channel Cav1.1 at 3.6 angstrom resolution
Wu, J.P., Yan, Z., Li, Z.Q., Qian, X.Y., Lu, S., Dong, M.Q., Zhou, Q., Yan, N.(2016) Nature 537: 191-196
- PubMed: 27580036
- DOI: https://doi.org/10.1038/nature19321
- Primary Citation of Related Structures:
5GJV - PubMed Abstract:
The voltage-gated calcium (Ca v ) channels convert membrane electrical signals to intracellular Ca 2+ -mediated events. Among the ten subtypes of Ca v channel in mammals, Ca v 1.1 is specified for the excitation-contraction coupling of skeletal muscles. Here we present the cryo-electron microscopy structure of the rabbit Ca v 1.1 complex at a nominal resolution of 3.6 Å. The inner gate of the ion-conducting α1-subunit is closed and all four voltage-sensing domains adopt an 'up' conformation, suggesting a potentially inactivated state. The extended extracellular loops of the pore domain, which are stabilized by multiple disulfide bonds, form a windowed dome above the selectivity filter. One side of the dome provides the docking site for the α2δ-1-subunit, while the other side may attract cations through its negative surface potential. The intracellular I-II and III-IV linker helices interact with the β 1a -subunit and the carboxy-terminal domain of α1, respectively. Classification of the particles yielded two additional reconstructions that reveal pronounced displacement of β 1a and adjacent elements in α1. The atomic model of the Ca v 1.1 complex establishes a foundation for mechanistic understanding of excitation-contraction coupling and provides a three-dimensional template for molecular interpretations of the functions and disease mechanisms of Ca v and Na v channels.
Organizational Affiliation:
State Key Laboratory of Membrane Biology, School of Life Sciences and School of Medicine, Tsinghua University, Beijing 100084, China.