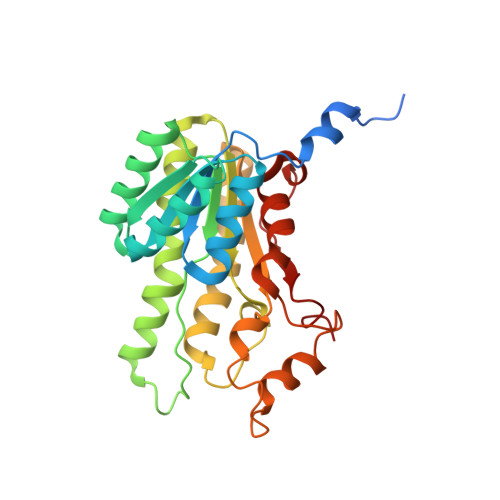
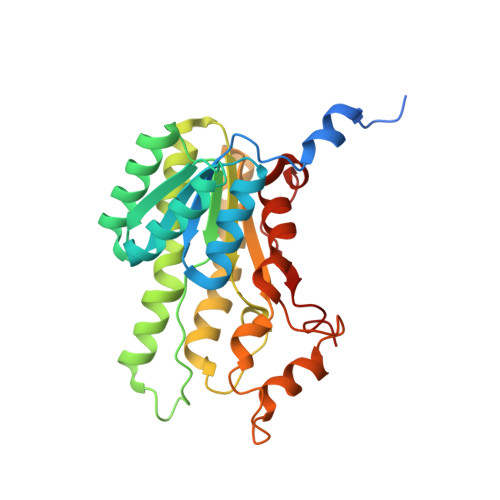
Biocatalytic Properties and Structural Analysis of Phloroglucinol Reductases.
Conradt, D., Hermann, B., Gerhardt, S., Einsle, O., Mueller, M.(2016) Angew Chem Int Ed Engl 55: 15531
- PubMed: 27874239
- DOI: https://doi.org/10.1002/anie.201607494
- Primary Citation of Related Structures:
5G4K, 5G4L - PubMed Abstract:
Phloroglucinol reductases (PGRs) are involved in anaerobic degradation in bacteria, in which they catalyze the dearomatization of phloroglucinol into dihydrophloroglucinol. We identified three PGRs, from different bacterial species, that are members of the family of NAD(P)H-dependent short-chain dehydrogenases/reductases (SDRs). In addition to catalyzing the reduction of the physiological substrate, the three enzymes exhibit activity towards 2,4,6-trihydroxybenzaldehyde, 2,4,6-trihydroxyacetophenone, and methyl 2,4,6-trihydroxybenzoate. Structural elucidation of PGRcl and comparison to known SDRs revealed a high degree of conservation. Several amino acid positions were identified as being conserved within the PGR subfamily and might be involved in substrate differentiation. The results enable the enzymatic dearomatization of monoaromatic phenol derivatives and provide insight into the functional diversity that may be found in families of enzymes displaying a high degree of structural homology.
Organizational Affiliation:
Institut für Pharmazeutische Wissenschaften, Albert-Ludwigs-Universität Freiburg, Albertstrasse 25, 79104, Freiburg, Germany.