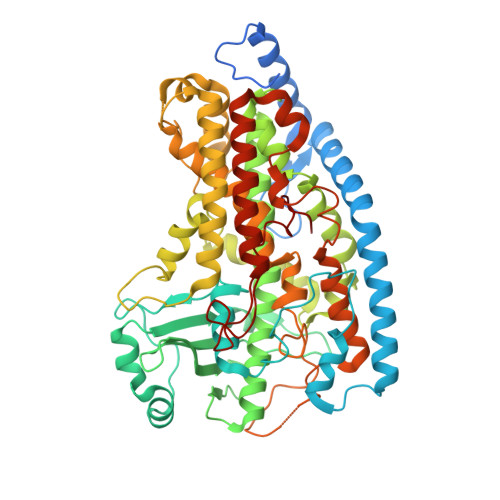
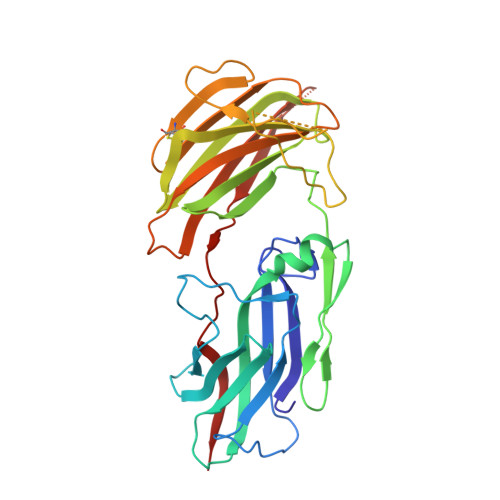
Crystal Structure of Linoleate 13R-Manganese Lipoxygenase and an Adhesion Protein
Chen, Y., Wennman, A., Karkehabadi, S., Engstrom, A., Oliw, E.H.(2016) J Lipid Res 57: 1574
- PubMed: 27313058
- DOI: https://doi.org/10.1194/jlr.M069617
- Primary Citation of Related Structures:
5FX8 - PubMed Abstract:
The crystal structure of 13R-manganese lipoxygenase (MnLOX) of Gaeumannomyces graminis (Gg) in complex with zonadhesin of Pichia pastoris was solved by molecular replacement. Zonadhesin contains β-strands in two subdomains. A comparison of Gg-MnLOX with the 9S-MnLOX of Magnaporthe oryzae (Mo) shows that the protein fold and the geometry of the metal ligands are conserved. The U-shaped active sites differ mainly due to hydrophobic residues of the substrate channel. The volumes and two hydrophobic side pockets near the catalytic base may sanction oxygenation at C-13 and C-9, respectively. Gly-332 of Gg-MnLOX is positioned in the substrate channel between the entrance and the metal center. Replacements with larger residues could restrict oxygen and substrate to reach the active site. C18 fatty acids are likely positioned with C-11 between Mn(2+)OH2 and Leu-336 for hydrogen abstraction and with one side of the 12Z double bond shielded by Phe-337 to prevent antarafacial oxygenation at C-13 and C-11. Phe-347 is positioned at the end of the substrate channel and replacement with smaller residues can position C18 fatty acids for oxygenation at C-9. Gg-MnLOX does not catalyze the sequential lipoxygenation of n-3 fatty acids in contrast to Mo-MnLOX, which illustrates the different configurations of their substrate channels.
Organizational Affiliation:
Department of Pharmaceutical Biosciences, Uppsala University Biomedical Center, SE-751 24 Uppsala, Sweden.