Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome.
Van den Bossche, A., Hardwick, S.W., Ceyssens, P.J., Hendrix, H., Voet, M., Dendooven, T., Bandyra, K.J., De Maeyer, M., Aertsen, A., Noben, J.P., Luisi, B.F., Lavigne, R.(2016) Elife 5
- PubMed: 27447594
- DOI: https://doi.org/10.7554/eLife.16413
- Primary Citation of Related Structures:
5FT0, 5FT1 - PubMed Abstract:
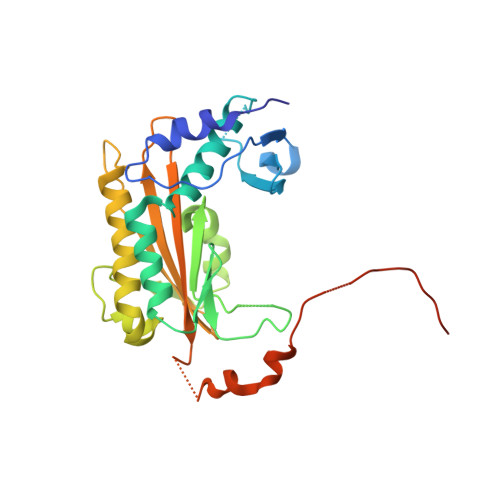
In all domains of life, the catalysed degradation of RNA facilitates rapid adaptation to changing environmental conditions, while destruction of foreign RNA is an important mechanism to prevent host infection. We have identified a virus-encoded protein termed gp37/Dip, which directly binds and inhibits the RNA degradation machinery of its bacterial host. Encoded by giant phage фKZ, this protein associates with two RNA binding sites of the RNase E component of the Pseudomonas aeruginosa RNA degradosome, occluding them from substrates and resulting in effective inhibition of RNA degradation and processing. The 2.2 Å crystal structure reveals that this novel homo-dimeric protein has no identifiable structural homologues. Our biochemical data indicate that acidic patches on the convex outer surface bind RNase E. Through the activity of Dip, фKZ has evolved a unique mechanism to down regulate a key metabolic process of its host to allow accumulation of viral RNA in infected cells.
Organizational Affiliation:
Laboratory of Gene Technology, KU Leuven, Leuven, Belgium.