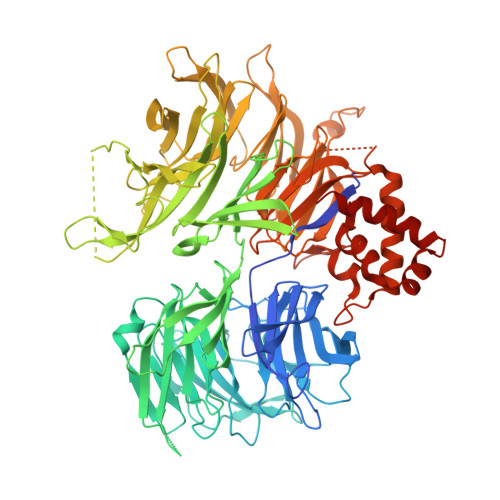
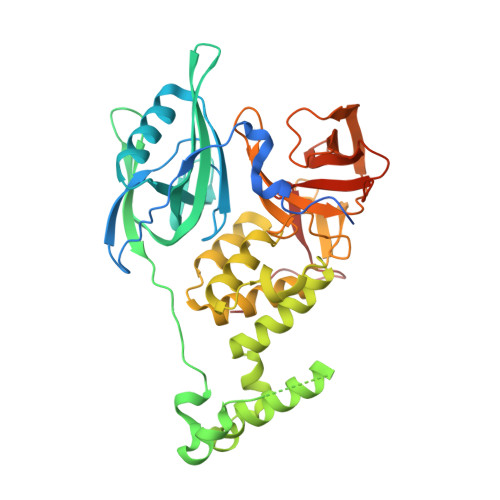
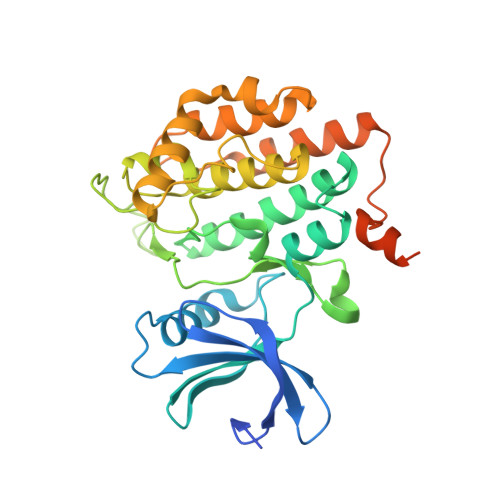
Structural Basis of Lenalidomide-Induced Ck1Alpha Degradation by the Crl4(Crbn) Ubiquitin Ligase.
Petzold, G., Fischer, E.S., Thoma, N.H.(2016) Nature 532: 127
- PubMed: 26909574
- DOI: https://doi.org/10.1038/nature16979
- Primary Citation of Related Structures:
5FQD - PubMed Abstract:
Thalidomide and its derivatives, lenalidomide and pomalidomide, are immune modulatory drugs (IMiDs) used in the treatment of haematologic malignancies. IMiDs bind CRBN, the substrate receptor of the CUL4-RBX1-DDB1-CRBN (also known as CRL4(CRBN)) E3 ubiquitin ligase, and inhibit ubiquitination of endogenous CRL4(CRBN) substrates. Unexpectedly, IMiDs also repurpose the ligase to target new proteins for degradation. Lenalidomide induces degradation of the lymphoid transcription factors Ikaros and Aiolos (also known as IKZF1 and IKZF3), and casein kinase 1α (CK1α), which contributes to its clinical efficacy in the treatment of multiple myeloma and 5q-deletion associated myelodysplastic syndrome (del(5q) MDS), respectively. How lenalidomide alters the specificity of the ligase to degrade these proteins remains elusive. Here we present the 2.45 Å crystal structure of DDB1-CRBN bound to lenalidomide and CK1α. CRBN and lenalidomide jointly provide the binding interface for a CK1α β-hairpin-loop located in the kinase N-lobe. We show that CK1α binding to CRL4(CRBN) is strictly dependent on the presence of an IMiD. Binding of IKZF1 to CRBN similarly requires the compound and both, IKZF1 and CK1α, use a related binding mode. Our study provides a mechanistic explanation for the selective efficacy of lenalidomide in del(5q) MDS therapy. We anticipate that high-affinity protein-protein interactions induced by small molecules will provide opportunities for drug development, particularly for targeted protein degradation.
Organizational Affiliation:
Friedrich Miescher Institute for Biomedical Research, Maulbeerstrasse 66, CH-4058 Basel, Switzerland.