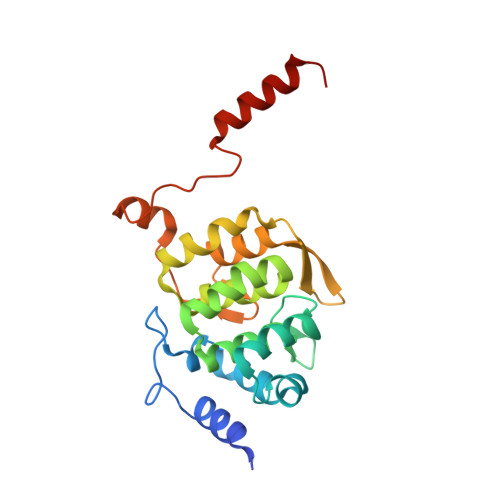
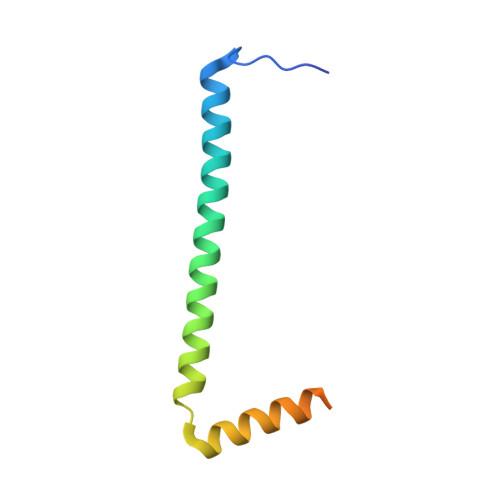
Scaffolding in the Spliceosome via Single alpha Helices.
Ulrich, A.K.C., Seeger, M., Schutze, T., Bartlick, N., Wahl, M.C.(2016) Structure 24: 1972-1983
- PubMed: 27773687
- DOI: https://doi.org/10.1016/j.str.2016.09.007
- Primary Citation of Related Structures:
5F5S, 5F5T, 5F5U, 5F5V - PubMed Abstract:
The spliceosomal B complex-specific protein Prp38 forms a complex with the intrinsically unstructured proteins MFAP1 and Snu23. Our binding and crystal structure analyses show that MFAP1 and Snu23 contact Prp38 via ER/K motif-stabilized single α helices, which have previously been recognized only as rigid connectors or force springs between protein domains. A variant of the Prp38-binding single α helix of MFAP1, in which ER/K motifs not involved in Prp38 binding were mutated, was less α-helical in isolation and showed a reduced Prp38 affinity, with opposing tendencies in interaction enthalpy and entropy. Our results indicate that the strengths of single α helix-based interactions can be tuned by the degree of helix stabilization in the unbound state. MFAP1, Snu23, and several other spliceosomal proteins contain multiple regions that likely form single α helices via which they might tether several binding partners and act as intermittent scaffolds that facilitate remodeling steps during assembly of an active spliceosome.
Organizational Affiliation:
Laboratory of Structural Biochemistry, Freie Universität Berlin, Takustraße 6, 14195 Berlin, Germany.