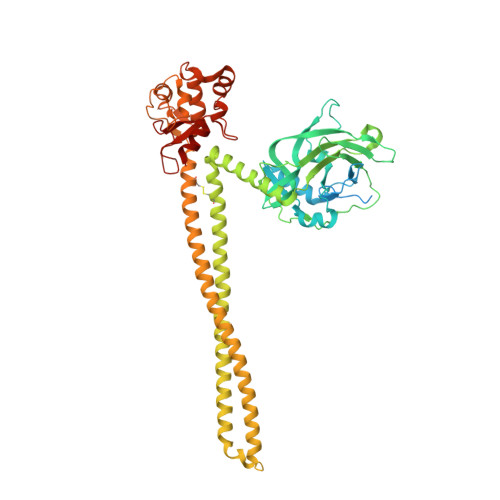
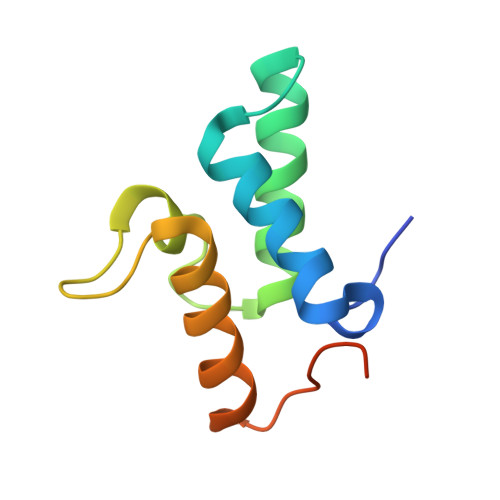
Structural and biophysical analysis of nuclease protein antibiotics.
Klein, A., Wojdyla, J.A., Joshi, A., Josts, I., McCaughey, L.C., Housden, N.G., Kaminska, R., Byron, O., Walker, D., Kleanthous, C.(2016) Biochem J 473: 2799-2812
- PubMed: 27402794
- DOI: https://doi.org/10.1042/BCJ20160544
- Primary Citation of Related Structures:
5EW5 - PubMed Abstract:
Protein antibiotics (bacteriocins) are a large and diverse family of multidomain toxins that kill specific Gram-negative bacteria during intraspecies competition for resources. Our understanding of the mechanism of import of such potent toxins has increased significantly in recent years, especially with the reporting of several structures of bacteriocin domains. Less well understood is the structural biochemistry of intact bacteriocins and how these compare across bacterial species. Here, we focus on endonuclease (DNase) bacteriocins that target the genomes of Escherichia coli and Pseudomonas aeruginosa, known as E-type colicins and S-type pyocins, respectively, bound to their specific immunity (Im) proteins. First, we report the 3.2 Å structure of the DNase colicin ColE9 in complex with its ultra-high affinity Im protein, Im9. In contrast with Im3, which when bound to the ribonuclease domain of the homologous colicin ColE3 makes contact with the translocation (T) domain of the toxin, we find that Im9 makes no such contact and only interactions with the ColE9 cytotoxic domain are observed. Second, we report small-angle X-ray scattering data for two S-type DNase pyocins, S2 and AP41, into which are fitted recently determined X-ray structures for isolated domains. We find that DNase pyocins and colicins are both highly elongated molecules, even though the order of their constituent domains differs. We discuss the implications of these architectural similarities and differences in the context of the translocation mechanism of protein antibiotics through the cell envelope of Gram-negative bacteria.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, U.K.