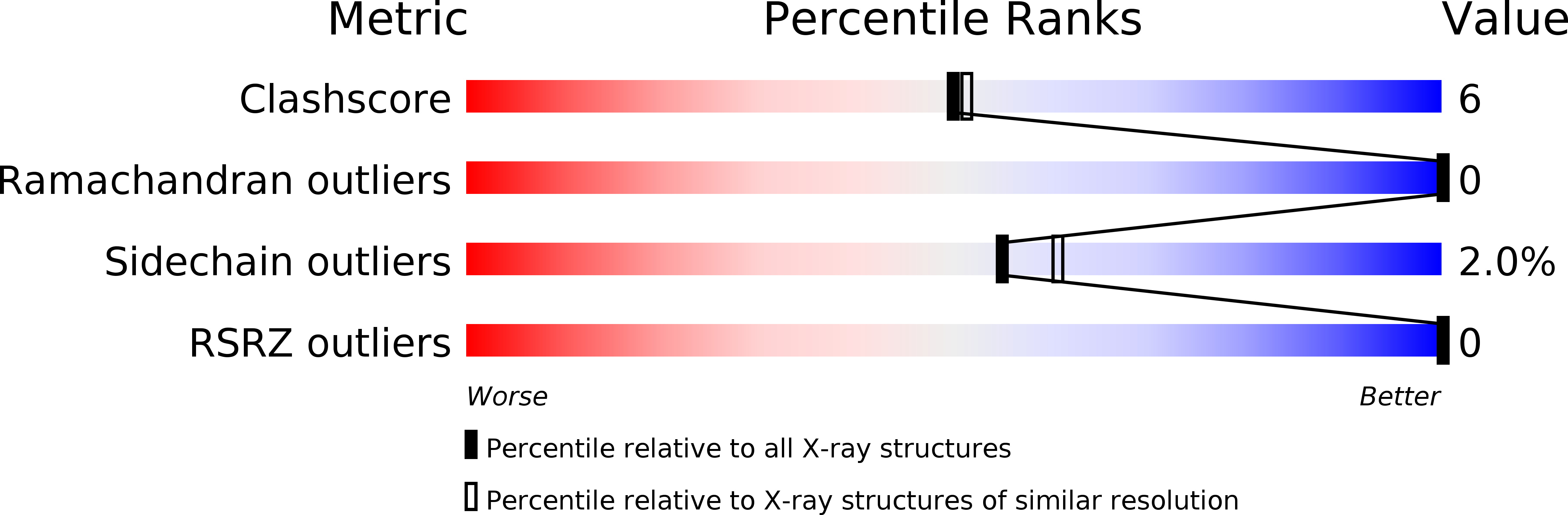
Crystallographic analysis of the inhibition of porcine pancreatic elastase by a peptidyl boronic acid: structure of a reaction intermediate.
Takahashi, L.H., Radhakrishnan, R., Rosenfield Jr., R.E., Meyer Jr., E.F.(1989) Biochemistry 28: 7610-7617
- PubMed: 2611205
- DOI: https://doi.org/10.1021/bi00445a016
- Primary Citation of Related Structures:
5EST - PubMed Abstract:
The crystal structure of porcine pancreatic elastase (PPE) complexed to carbobenzoxy-alanylisoleucine-boronic acid (ZAIB) is reported to 2.09-A resolution and refined to an R factor of 0.15. This is the first reported structural analysis of PPE with an isoleucine residu in the primary specificity pocket. The results include (1) marked displacement of the inhibitor out of the active site leading to (2) a close (2.2 A) direct contact between B (boron atom of the inhibitor) and N epsilon of His-57 and also (3) covalent bonding (1.5 A) to O gamma of Ser-195. A scheme for the mechanism of inhibition of PPE by ZAIB is proposed. A comparison with a peptidyl difluoromethyl ketone-PPE complex (Ki = 9.5 microns) is made to explain the strong inhibition of PPE by ZAIB (Ki = 0.3 micron). These results lead us to characterize this structure as a time- and space-averaged reaction intermediate, providing fresh insight into the cramped dimensions available in enzymatic catalyses.
Organizational Affiliation:
Department of Chemistry, Texas A&M University, College Station 77843.