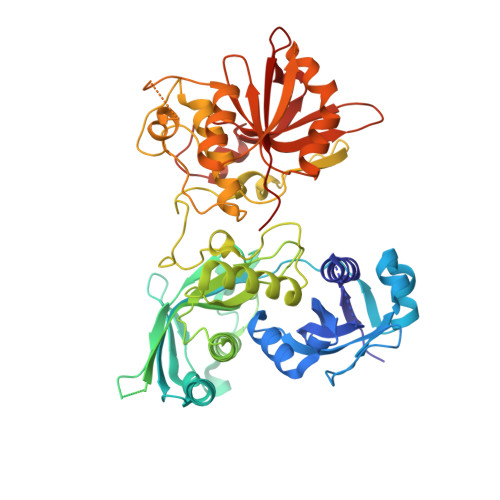
The MPN domain of Caenorhabditis elegans UfSP modulates both substrate recognition and deufmylation activity
Ha, B.H., Kim, K.H., Yoo, H.M., Lee, W., Kim, E.E.(2016) Biochem Biophys Res Commun 476: 450-456
- PubMed: 27240952
- DOI: https://doi.org/10.1016/j.bbrc.2016.05.143
- Primary Citation of Related Structures:
5EJJ - PubMed Abstract:
Ubiquitin-fold modifier 1 (Ufm1) specific protease (UfSP) is a novel cysteine protease that activates Ufm1 from its precursor by processing the C-terminus to expose the conserved Gly necessary for substrate conjugation and de-conjugates Ufm1 from the substrate. There are two forms: UfSP1 and UfSP2, the later with an additional domain at the N-terminus. Ufm1 and both the conjugating and deconjugating enzymes are highly conserved. However, in Caenorhabditis elegans there is one UfSP which has extra 136 residues at the N terminus compared to UfSP2. The crystal structure of cUfSP reveals that these additional residues display a MPN fold while the rest of the structure mimics that of UfSP2. The MPN domain does not have the metalloprotease activity found in some MPN-domain containing protein, rather it is required for the recognition and deufmylation of the substrate of cUfSP, UfBP1. In addition, the MPN domain is also required for localization to the endoplasmic reticulum.
Organizational Affiliation:
Biomedical Research Institute, Korea Institute of Science and Technology, Seoul 136-791, Republic of Korea.