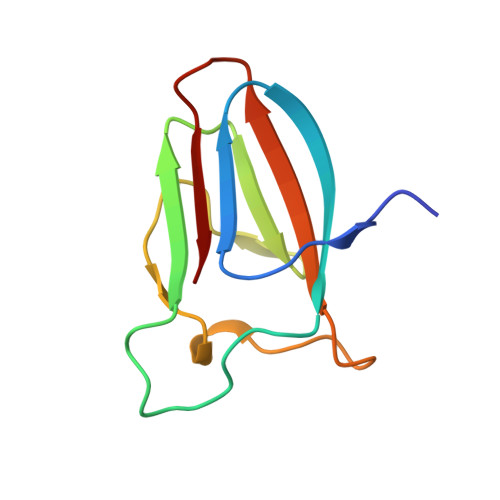
Structural basis for cellulose binding by the type A carbohydrate-binding module 64 of Spirochaeta thermophila.
Schiefner, A., Angelov, A., Liebl, W., Skerra, A.(2016) Proteins 84: 855-858
- PubMed: 26868291
- DOI: https://doi.org/10.1002/prot.25010
- Primary Citation of Related Structures:
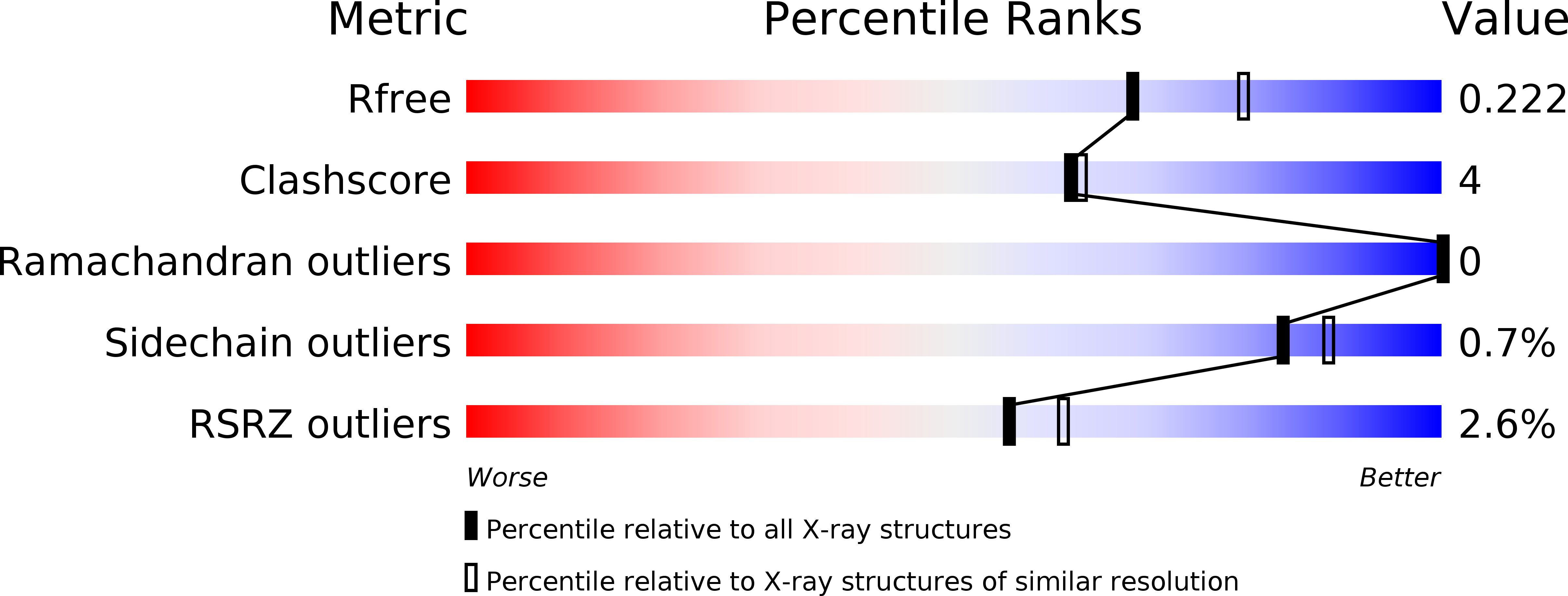
5E9O, 5E9P - PubMed Abstract:
Spirochaeta thermophila secretes seven glycoside hydrolases for plant biomass degradation that carry a carbohydrate-binding module 64 (CBM64) appended at the C-terminus. CBM64 adsorbs to various β1-4-linked pyranose substrates and shows high affinity for cellulose. We present the first crystal structure of a CBM64 at 1.2 Å resolution, which reveals a jelly-roll-like fold corresponding to a surface-binding type A CBM. Modeling of its interaction with cellulose indicates that CBM64 achieves association with the hydrophobic face of β-linked pyranose chains via a unique coplanar arrangement of four exposed tryptophan side chains. Proteins 2016; 84:855-858. © 2016 Wiley Periodicals, Inc.
Organizational Affiliation:
Munich Center for Integrated Protein Science (CIPS-M) and Lehrstuhl für Biologische Chemie, Technische Universität München, 85354 Freising (Weihenstephan), Germany.