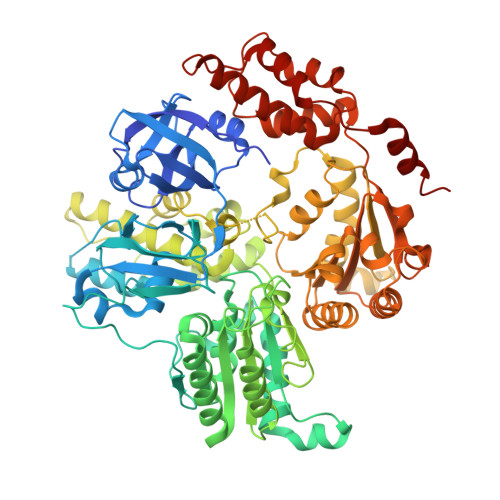
Crystal Structure and Biochemical Characterization of a Mycobacterium smegmatis AAA-Type Nucleoside Triphosphatase Phosphohydrolase (Msm0858).
Unciuleac, M.C., Smith, P.C., Shuman, S.(2016) J Bacteriol 198: 1521-1533
- PubMed: 26953339
- DOI: https://doi.org/10.1128/JB.00905-15
- Primary Citation of Related Structures:
5E7P - PubMed Abstract:
AAA proteins (ATPases associated with various cellular activities) use the energy of ATP hydrolysis to drive conformational changes in diverse macromolecular targets. Here, we report the biochemical characterization and 2.5-Å crystal structure of a Mycobacterium smegmatis AAA protein Msm0858, the ortholog of Mycobacterium tuberculosis Rv0435c. Msm0858 is a magnesium-dependent ATPase and is active with all nucleoside triphosphates (NTPs) and deoxynucleoside triphosphates (dNTPs) as substrates. The Msm0858 structure comprises (i) an N-terminal domain (amino acids [aa] 17 to 201) composed of two β-barrel modules and (ii) two AAA domains, D1 (aa 212 to 473) and D2 (aa 476 to 744), each of which has ADP in the active site. Msm0858-ADP is a monomer in solution and in crystallized form. Msm0858 domains are structurally homologous to the corresponding modules of mammalian p97. However, the position of the N-domain modules relative to the AAA domains in the Msm0858-ADP tertiary structure is different and would impede the formation of a p97-like hexameric quaternary structure. Mutational analysis of the A-box and B-box motifs indicated that the D1 and D2 AAA domains are both capable of ATP hydrolysis. Simultaneous mutations of the D1 and D2 active-site motifs were required to abolish ATPase activity. ATPase activity was effaced by mutation of the putative D2 arginine finger, suggesting that Msm0858 might oligomerize during the ATPase reaction cycle. A truncated variant Msm0858 (aa 212 to 745) that lacks the N domain was characterized as a catalytically active homodimer.
Organizational Affiliation:
Molecular Biology Program, Sloan-Kettering Institute, New York, New York, USA.