Identification and Characterization of a New Pecan [Carya illinoinensis (Wangenh.) K. Koch] Allergen, Car i 2.
Zhang, Y., Lee, B., Du, W.X., Lyu, S.C., Nadeau, K.C., Grauke, L.J., Zhang, Y., Wang, S., Fan, Y., Yi, J., McHugh, T.H.(2016) J Agric Food Chem 64: 4146-4151
- PubMed: 27128197
- DOI: https://doi.org/10.1021/acs.jafc.6b00884
- Primary Citation of Related Structures:
5E1R - PubMed Abstract:
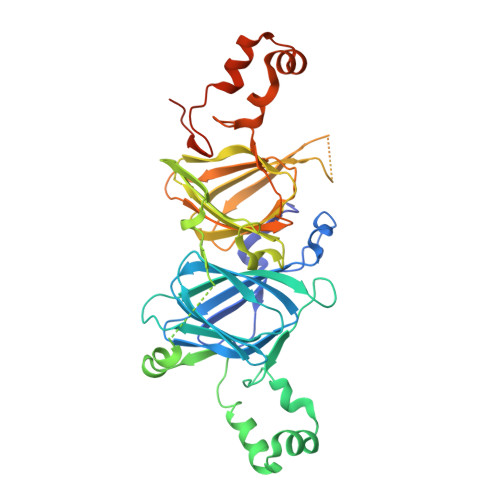
The 7S vicilin and 11S legumin seed storage globulins belong to the cupin protein superfamily and are major food allergens in many foods from the "big eight" food allergen groups. Here, for the first time, pecan vicilin was found to be a food allergen. Western blot experiments revealed that 30% of 27 sera used in this study and 24% of the sera from 25 patients with double-blind, placebo controlled clinical pecan allergy contained IgE antibodies specific to pecan vicilin. This allergen consists of a low-complexity region at its N-terminal and a structured domain at the C-terminal that contains two cupin motifs and forms homotrimers. The crystal structure of recombinant pecan vicilin was determined. The refined structure gave R/Rfree values of 0.218/0.262 for all data to 2.65 Å. There were two trimeric biological units in the crystallographic asymmetric unit. Pecan vicilin is also a copper protein. These data may facilitate the understanding of the nutritional value and the allergenicity relevance of the copper binding property of seed storage proteins in tree nuts.
Organizational Affiliation:
Western Regional Research Center, Pacific West Area, Agricultural Research Service, U.S. Department of Agriculture , 800 Buchanan Street, Albany, California 94710, United States.