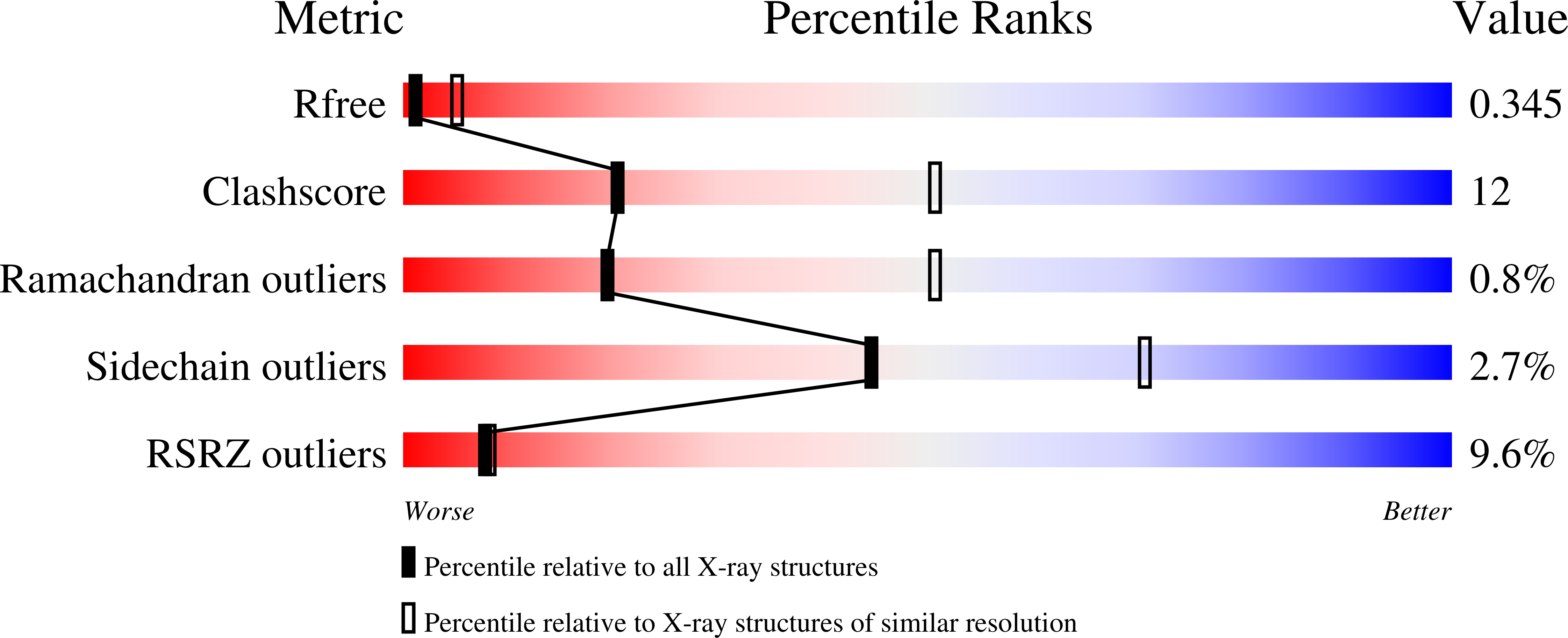
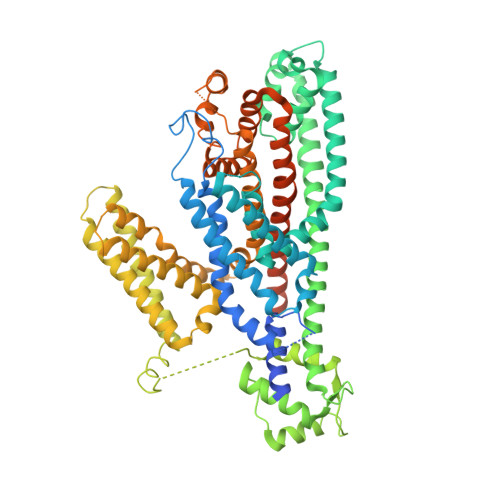
Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana.
Guo, J., Zeng, W., Chen, Q., Lee, C., Chen, L., Yang, Y., Cang, C., Ren, D., Jiang, Y.(2016) Nature 531: 196-201
- PubMed: 26689363
- DOI: https://doi.org/10.1038/nature16446
- Primary Citation of Related Structures:
5E1J - PubMed Abstract:
Two-pore channels (TPCs) contain two copies of a Shaker-like six-transmembrane (6-TM) domain in each subunit and are ubiquitously expressed in both animals and plants as organellar cation channels. Here we present the crystal structure of a vacuolar two-pore channel from Arabidopsis thaliana, AtTPC1, which functions as a homodimer. AtTPC1 activation requires both voltage and cytosolic Ca(2+). Ca(2+) binding to the cytosolic EF-hand domain triggers conformational changes coupled to the pair of pore-lining inner helices from the first 6-TM domains, whereas membrane potential only activates the second voltage-sensing domain, the conformational changes of which are coupled to the pair of inner helices from the second 6-TM domains. Luminal Ca(2+) or Ba(2+) can modulate voltage activation by stabilizing the second voltage-sensing domain in the resting state and shift voltage activation towards more positive potentials. Our Ba(2+)-bound AtTPC1 structure reveals a voltage sensor in the resting state, providing hitherto unseen structural insight into the general voltage-gating mechanism among voltage-gated channels.
Organizational Affiliation:
Department of Physiology, University of Texas Southwestern Medical Center, Dallas, Texas 75390-9040, USA.