A Dye-Decolorizing Peroxidase from Vibrio cholerae.
Uchida, T., Sasaki, M., Tanaka, Y., Ishimori, K.(2015) Biochemistry 54: 6610-6621
- PubMed: 26431465
- DOI: https://doi.org/10.1021/acs.biochem.5b00952
- Primary Citation of Related Structures:
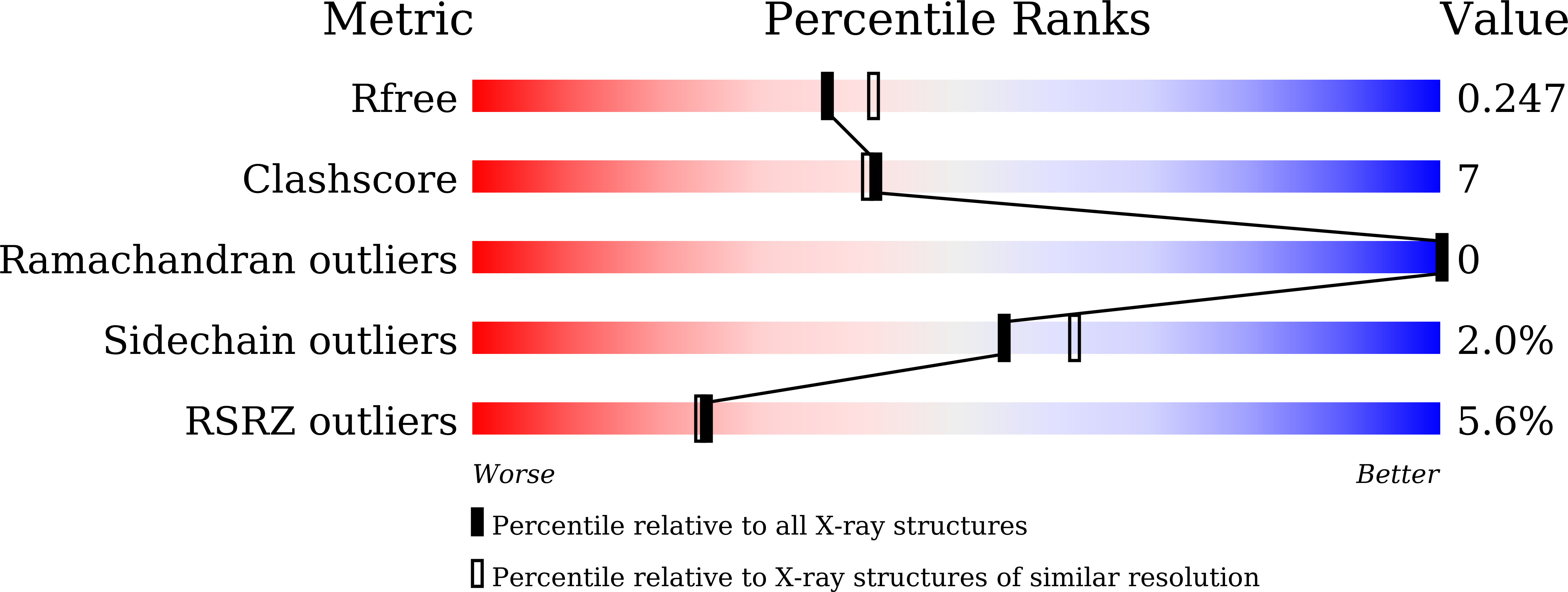
5DE0 - PubMed Abstract:
The dye-decolorizing peroxidase (DyP) protein from Vibrio cholerae (VcDyP) was expressed in Escherichia coli, and its DyP activity was assayed by monitoring degradation of a typical anthraquinone dye, reactive blue 19 (RB19). Its kinetic activity was obtained by fitting the data to the Michaelis-Menten equation, giving kcat and Km values of 1.3 ± 0.3 s(-1) and 50 ± 20 μM, respectively, which are comparable to those of other DyP enzymes. The enzymatic activity of VcDyP was highest at pH 4. A mutational study showed that two distal residues, Asp144 and Arg230, which are conserved in a DyP family, are essential for the DyP reaction. The crystal structure and resonance Raman spectra of VcDyP indicate the transfer of a radical from heme to the protein surface, which was supported by the formation of the intermolecular covalent bond in the reaction with H2O2. To identify the radical site, each of nine tyrosine or two tryptophan residues was substituted. It was clarified that Tyr129 and Tyr235 are in the active site of the dye degradation reaction at lower pH, while Tyr109 and Tyr133 are the sites of an intermolecular covalent bond at higher pH. VcDyP degrades RB19 at lower pH, while it loses activity under neutral or alkaline conditions because of a change in the radical transfer pathway. This finding suggests the presence of a pH-dependent switch of the radical transfer pathway, probably including His178. Although the physiological function of the DyP reaction is unclear, our findings suggest that VcDyP enhances the DyP activity to survive only when it is placed under a severe condition such as being in gastric acid.
Organizational Affiliation:
Department of Chemistry, Faculty of Science, Hokkaido University , Sapporo 060-0810, Japan.