Structure of a thermostable serralysin from Serratia sp. FS14 at 1.1 angstrom resolution.
Wu, D., Ran, T., Wang, W., Xu, D.(2016) Acta Crystallogr F Struct Biol Commun 72: 10-15
- PubMed: 26750478
- DOI: https://doi.org/10.1107/S2053230X15023092
- Primary Citation of Related Structures:
5D7W - PubMed Abstract:
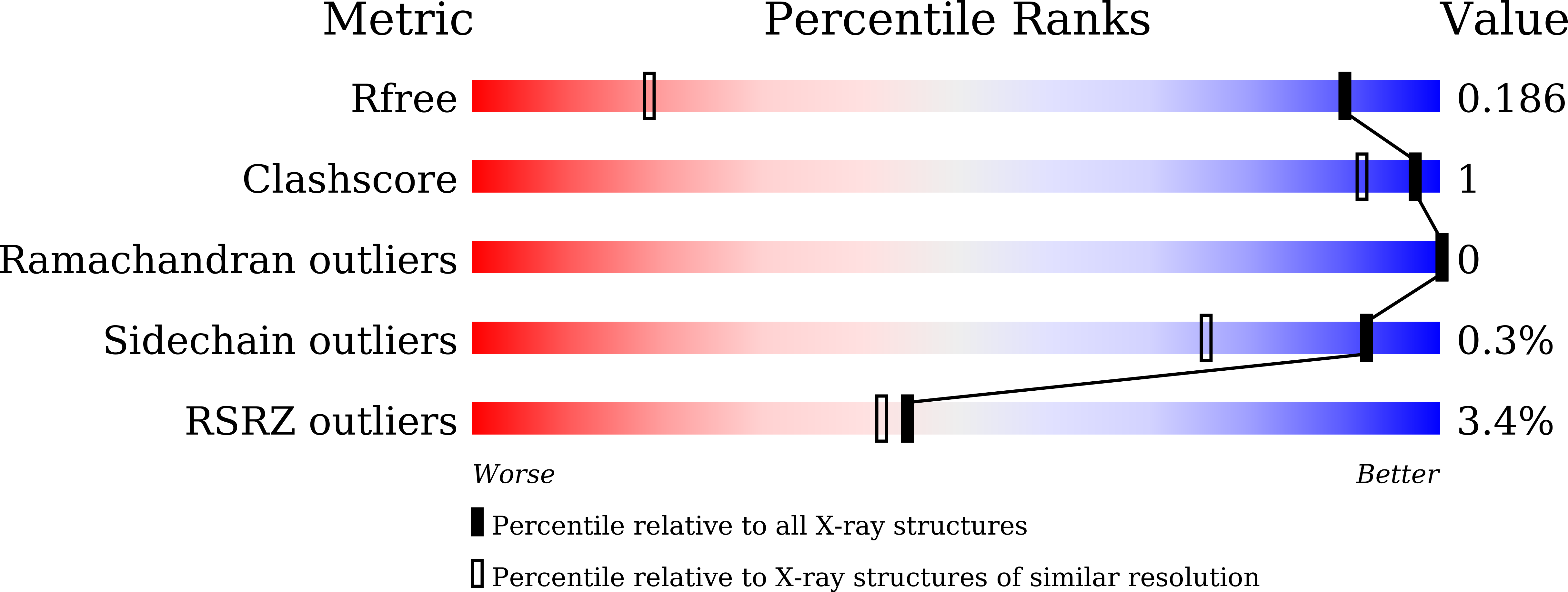
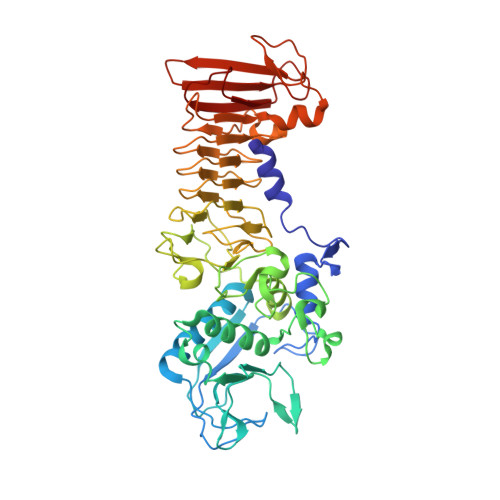
Serralysin is a well studied metalloprotease, and typical serralysins are not thermostable. The serralysin isolated from Serratia sp. FS14 was found to be thermostable, and in order to reveal the mechanism responsible for its thermostability, the crystal structure of serralysin from Serratia sp. FS14 was solved to a crystallographic R factor of 0.1619 at 1.10 Å resolution. Similar to its homologues, it mainly consists of two domains: an N-terminal catalytic domain and a `parallel β-roll' C-terminal domain. Comparative studies show that the shape of the catalytic active-site cavity is more open owing to the 189-198 loop, with a short 310-helix protruding further from the molecular surface, and that the β-sheets comprising the `parallel β-roll' are longer than those in its homologues. The formation of hydrogen bonds from one of the nonconserved residues (Asn200) to Lys27 may contribute to the thermostability.
Organizational Affiliation:
Department of Microbiology, Nanjing Agricultural University, No. 1 Weigang, Nanjing, Jiangsu 210095, People's Republic of China.