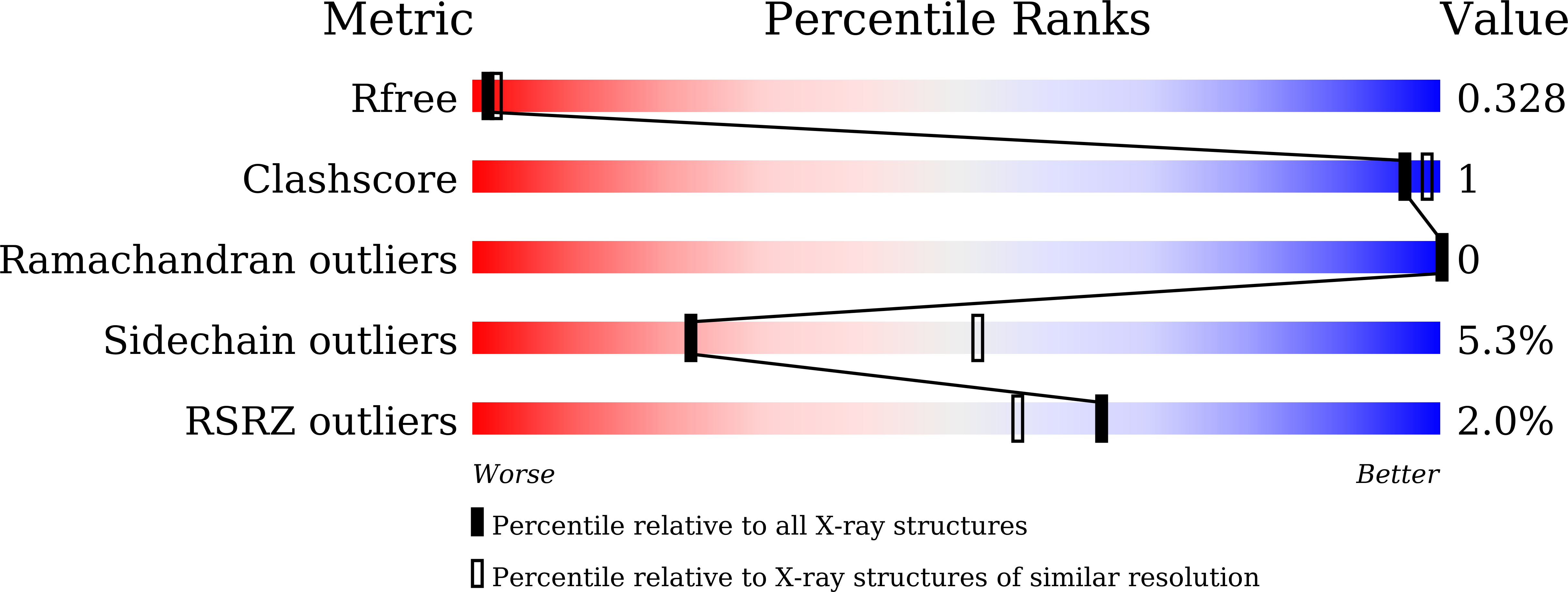
Crystal Structure of Wild-Type Centrin 1 from Mus musculus Occupied by Ca2.
Kim, S.Y., Kim, D.S., Hong, J.E., Park, J.H.(2017) Biochemistry (Mosc) 82: 1129-1139
- PubMed: 29037133
- DOI: https://doi.org/10.1134/S0006297917100054
- Primary Citation of Related Structures:
5D43 - PubMed Abstract:
Mus musculus centrin 1 (MmCen1) is located at the cilium of photoreceptor cells connecting the outer segment through signal transduction components to the metabolically active inner segment. In the cilium, MmCen1 is involved in the translocation of transducin between compartments as a result of photoreceptor activation. In this study, we report the crystal structure of wild-type MmCen1 and its Ca2+-binding properties using structure-based mutagenesis. The crystal structure exhibits three structural features, i.e. four Ca2+ equally occupied at each EF-hand motif, structural changes accompanying helix motion at the N- and C-lobes, and adoption of N-C type dimerization when Ca2+ binds to EF-hand I and II in the N-lobe. The presence of MmCen1 dimers was confirmed in solution by native PAGE. Isothermal titration calorimetry data showed sequential binding of Ca2+ at four independent sites. Mutations S45A and D49A in EF-hand I alone disrupted the Ca2+-binding property of the wild-type protein. Based on the crystal structure of MmCen1, we suggest that a dimerization mode between the N- and C-lobes may be required by Ca2+ binding at the N-lobe.
Organizational Affiliation:
Chonbuk National University, College of Environmental & Bioresource Sciences, Division of Biotechnology, Iksan, 54596, Republic of Korea. junghee.park@jbnu.ac.kr.