Tubulin cofactors and Arl2 are cage-like chaperones that regulate the soluble alpha beta-tubulin pool for microtubule dynamics.
Nithianantham, S., Le, S., Seto, E., Jia, W., Leary, J., Corbett, K.D., Moore, J.K., Al-Bassam, J.(2015) Elife 4
- PubMed: 26208336
- DOI: https://doi.org/10.7554/eLife.08811
- Primary Citation of Related Structures:
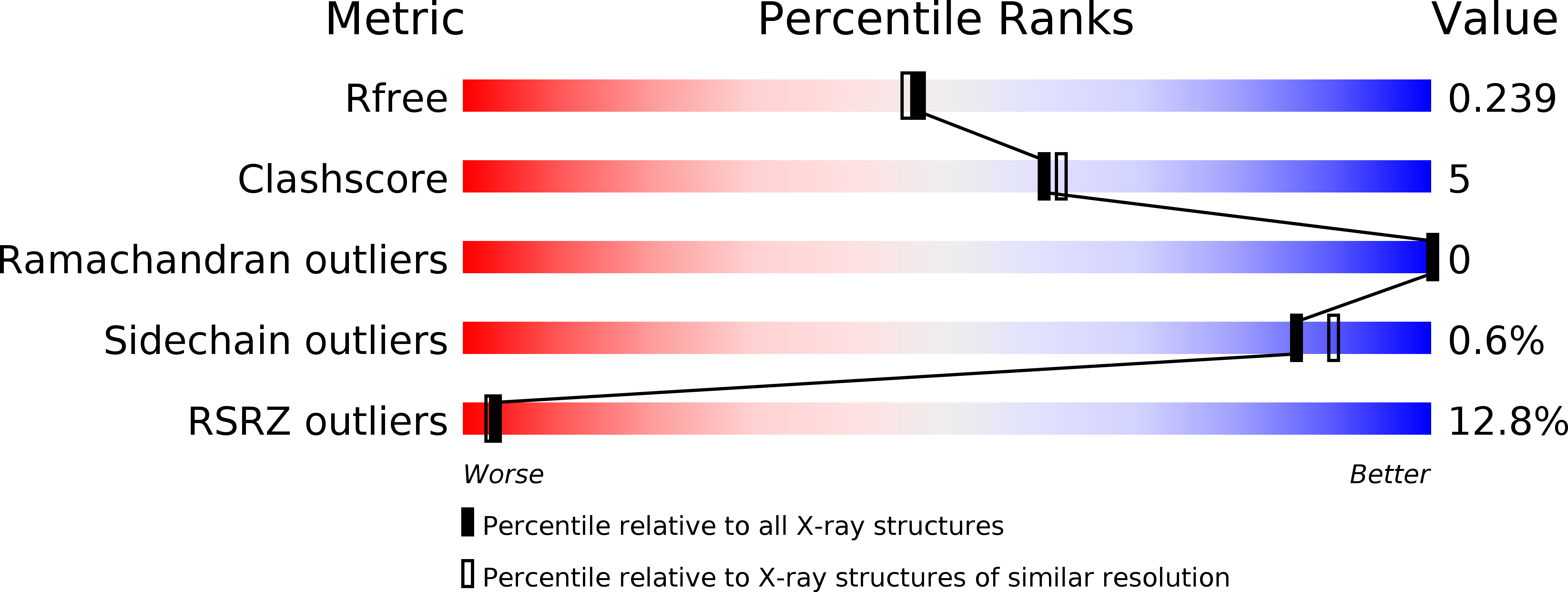
5CYA - PubMed Abstract:
Microtubule dynamics and polarity stem from the polymerization of αβ-tubulin heterodimers. Five conserved tubulin cofactors/chaperones and the Arl2 GTPase regulate α- and β-tubulin assembly into heterodimers and maintain the soluble tubulin pool in the cytoplasm, but their physical mechanisms are unknown. Here, we reconstitute a core tubulin chaperone consisting of tubulin cofactors TBCD, TBCE, and Arl2, and reveal a cage-like structure for regulating αβ-tubulin. Biochemical assays and electron microscopy structures of multiple intermediates show the sequential binding of αβ-tubulin dimer followed by tubulin cofactor TBCC onto this chaperone, forming a ternary complex in which Arl2 GTP hydrolysis is activated to alter αβ-tubulin conformation. A GTP-state locked Arl2 mutant inhibits ternary complex dissociation in vitro and causes severe defects in microtubule dynamics in vivo. Our studies suggest a revised paradigm for tubulin cofactors and Arl2 functions as a catalytic chaperone that regulates soluble αβ-tubulin assembly and maintenance to support microtubule dynamics.
Organizational Affiliation:
Department of Molecular Cellular Biology, University of California, Davis, Davis, United States.