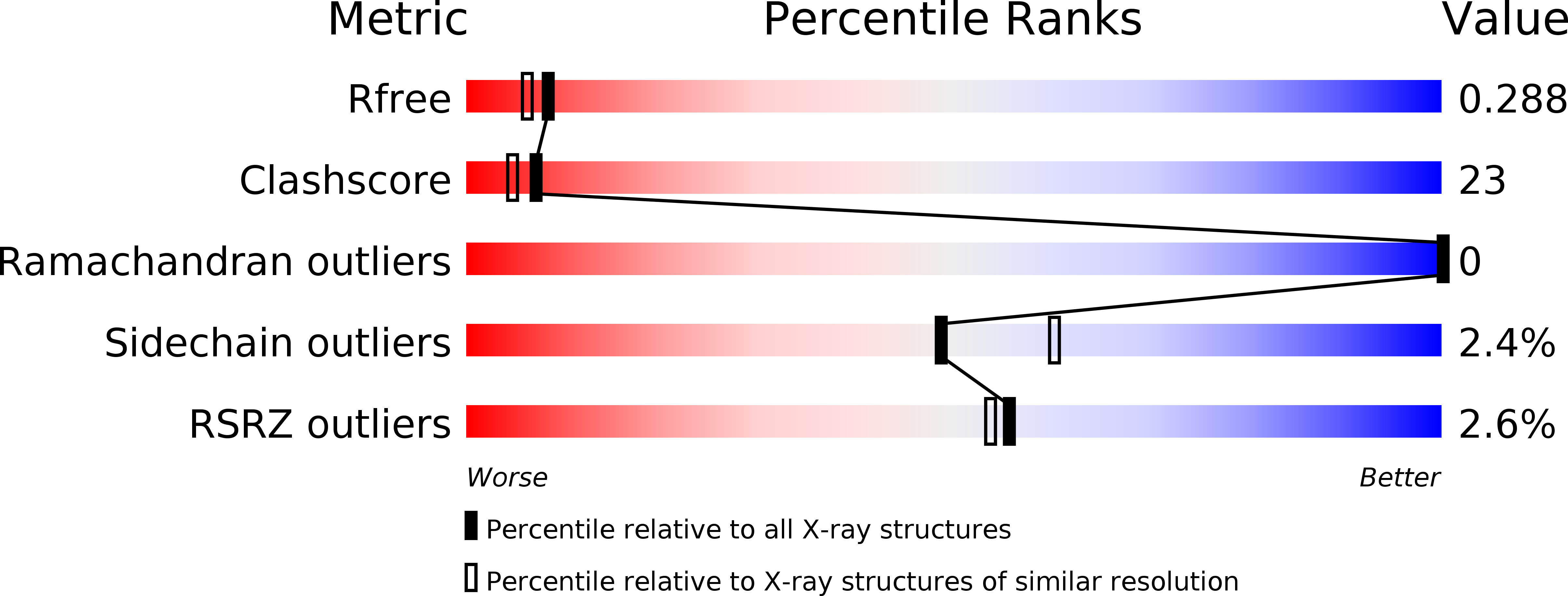
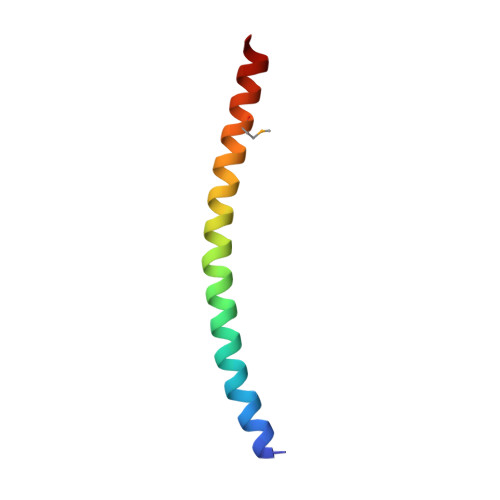Structure of Leishmania donovani coronin coiled coil domain reveals an antiparallel 4 helix bundle with inherent asymmetry
Nayak, A.R., Karade, S.S., Srivastava, V.K., Rana, A.K., Gupta, C.M., Sahasrabuddhe, A.A., Pratap, J.V.(2016) J Struct Biol 195: 129-138
- PubMed: 26940672
- DOI: https://doi.org/10.1016/j.jsb.2016.02.020
- Primary Citation of Related Structures:
5CX2 - PubMed Abstract:
Coiled coils are ubiquitous structural motifs that serve as a platform for protein-protein interactions and play a central role in myriad physiological processes. Though the formation of a coiled coil requires only the presence of suitably spaced hydrophobic residues, sequence specificities have also been associated with specific oligomeric states. RhXXhE is one such sequence motif, associated with parallel trimers, found in coronins and other proteins. Coronin, present in all eukaryotes, is an actin-associated protein involved in regulating actin turnover. Most eukaryotic coronins possess the RhXXhE trimerization motif. However, a unique feature of parasitic kinetoplastid coronin is that the positions of R and E are swapped within their coiled coil domain, but were still expected to form trimers. To understand the role of swapped motif in oligomeric specificity, we determined the X-ray crystal structure of Leishmania donovani coronin coiled coil domain (LdCoroCC) at 2.2Å, which surprisingly, reveals an anti-parallel tetramer assembly. Small angle X-ray scattering studies and chemical crosslinking confirm the tetramer in solution and is consistent with the oligomerization observed in the full length protein. Structural analyses reveal that LdCoroCC possesses an inherent asymmetry, in that one of the helices of the bundle is axially shifted with respect to the other three. The analysis also identifies steric reasons that cause this asymmetry. The bundle adapts an extended a-d-e core packing, the e residue being polar (with an exception) which results in a thermostable bundle with polar and apolar interfaces, unlike the existing a-d-e core antiparallel homotetramers with apolar core. Functional implications of the anti-parallel association in kinetoplastids are discussed.
Organizational Affiliation:
Division of Molecular and Structural Biology, CSIR - Central Drug Research Institute, Jankipuram Extension, Sitapur Road, Lucknow 226031, India.