Crystal structure of maize serine racemase with pyridoxal 5'-phosphate.
Zou, L., Song, Y., Wang, C., Sun, J., Wang, L., Cheng, B., Fan, J.(2016) Acta Crystallogr F Struct Biol Commun 72: 165-171
- PubMed: 26919519
- DOI: https://doi.org/10.1107/S2053230X16000960
- Primary Citation of Related Structures:
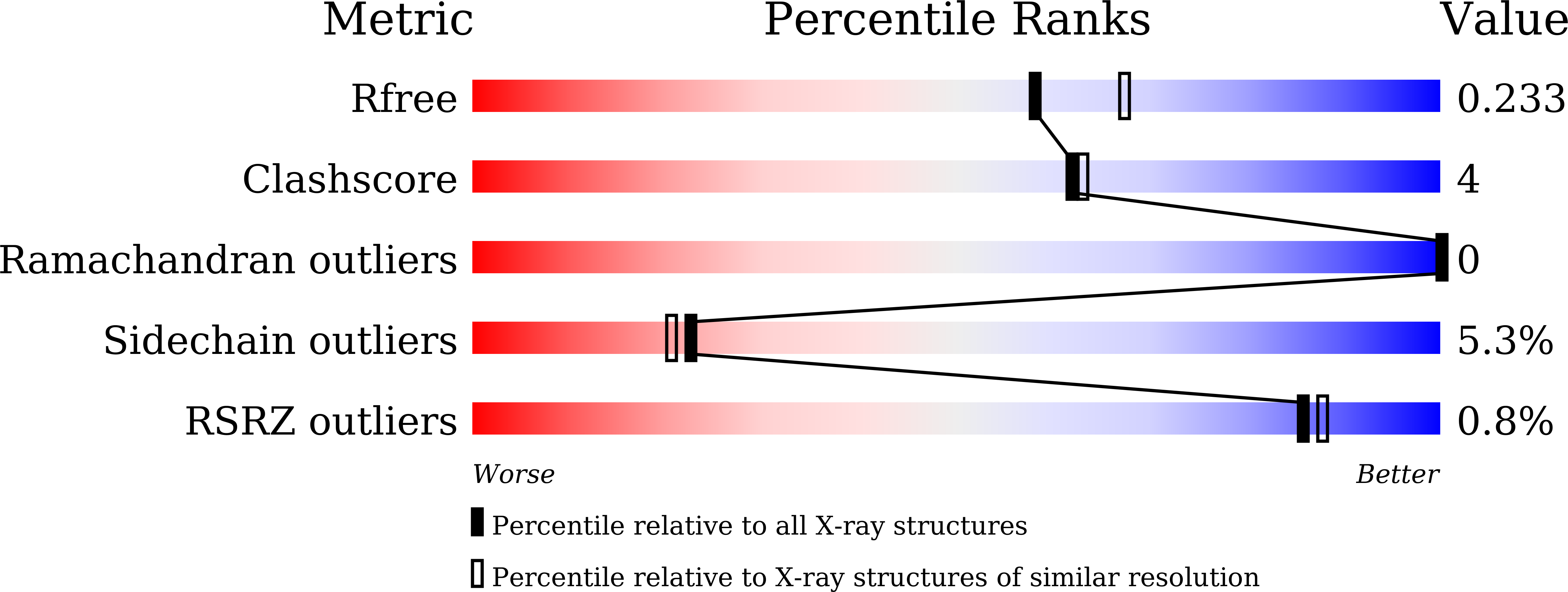
5CVC - PubMed Abstract:
Serine racemase (SR) is a pyridoxal 5'-phosphate (PLP)-dependent enzyme that is responsible for D-serine biosynthesis in vivo. The first X-ray crystal structure of maize SR was determined to 2.1 Å resolution and PLP binding was confirmed in solution by UV-Vis absorption spectrometry. Maize SR belongs to the type II PLP-dependent enzymes and differs from the SR of a vancomycin-resistant bacterium. The PLP is bound to each monomer by forming a Schiff base with Lys67. Structural comparison with rat and fission yeast SRs reveals a similar arrangement of active-site residues but a different orientation of the C-terminal helix.
Organizational Affiliation:
School of Life Science, Anhui Agricultural University, Hefei, Anhui 230036, People's Republic of China.