Mechanistic insight into mucin degradation by the human gut microbiota reveals targeting of the glycoprotein core
Ndeh, D.A., Nakjang, S., Kwiatkowski, K.J., Koropatkin, N.M., Hirt, R.P., Bolam, D.N.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| SusD homolog | 641 | Bacteroides thetaiotaomicron VPI-5482 | Mutation(s): 0 Gene Names: BT_4246 | 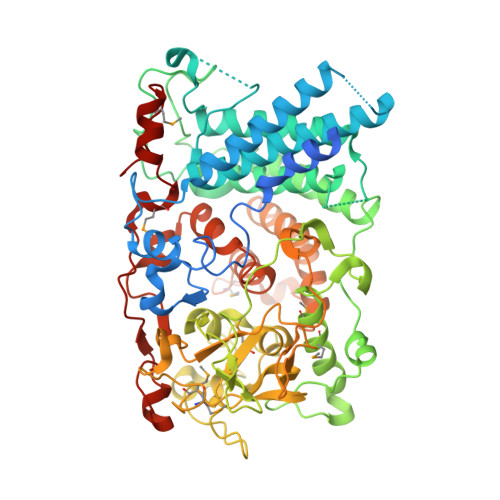 | |
UniProt | |||||
Find proteins for Q89ZX5 (Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50)) Explore Q89ZX5 Go to UniProtKB: Q89ZX5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q89ZX5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EDO Query on EDO | B [auth A], C [auth A], D [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CA Query on CA | E [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 110.792 | α = 90 |
| b = 115.738 | β = 90 |
| c = 116.656 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| University of Michigan Host Microbiome Initiative | United States | -- |