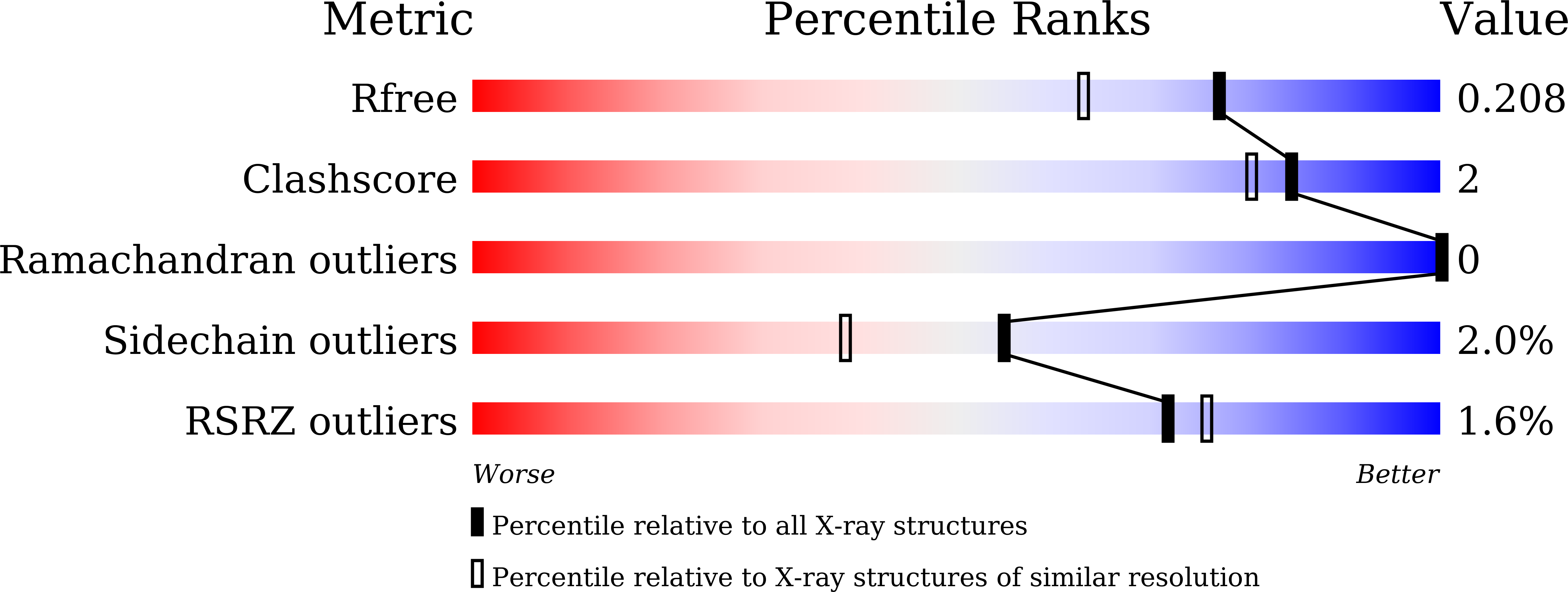
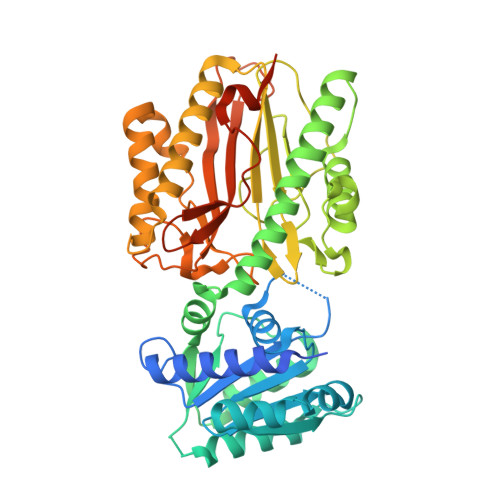
Crystal structure and dynamics of Spt16N-domain of FACT complex from Cicer arietinum.
Are, V.N., Ghosh, B., Kumar, A., Gadre, R., Makde, R.D.(2016) Int J Biol Macromol 88: 36-43
- PubMed: 26995613
- DOI: https://doi.org/10.1016/j.ijbiomac.2016.03.029
- Primary Citation of Related Structures:
5CE6 - PubMed Abstract:
The facilitates chromatin transcription (FACT) complex, a heterodimer of SSRP1 and Spt16 proteins, is an essential histone chaperone that transiently reorganizes nucleosomes during transcription, replication and repair. N-terminal domain of Spt16 subunit (Spt16N) is strictly conserved in all the known Spt16 orthologs. Genetic studies in yeast have revealed a partially redundant role of Spt16N for the FACT functionality. Here, we report the crystal structure of Spt16N from a plant origin (Spt16Nca, Cicer arietinum) and its comparisons with the known Spt16N structures from yeasts and human. The inter-domain angle in Spt16Nca is significantly different from that of the yeast and human Spt16N structures. Normal mode analysis and classical molecular dynamics simulations reveal inter-domain movement in Spt16Nca and later also shows conformational flexibility of the critical loops. Spt16Nca binds to histone H3/H4 complex, similar to its orthologs from yeast and human origins. Further, conservation of electrostatic surface potentials in Spt16N structures from evolutionary distinct domains of eukaryotes (plant, human and fungi) have provided the potential sites on Spt16N for histone interactions. The structural comparisons with M24 peptidases show that the hydrophobic pocket shielded by a flexible loop of C-terminal domain of Spt16N that may be functionally important.
Organizational Affiliation:
High Pressure and Synchrotron Radiation Physics Division, Bhabha Atomic Research Centre, Mumbai, India; School of Biochemistry, Devi Ahilya University, Indore, India.