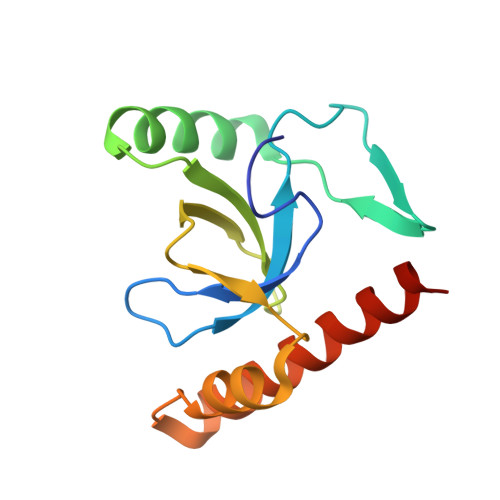
Human Bromodomain and PHD Finger Containing 1, PWWP domain in complex with XST005904a
DOBROVETSKY, E., DONG, A., MADER, P., Santhakumar, S., TEMPEL, W., Arrowsmith, C.H., Edwards, A.M., BROWN, P.J., Structural Genomics Consortium (SGC)To be published.