Structure of Mcl-1 complexed with Mule at 2.05 Angstroms resolution
Song, T., Wang, Z., Ji, F., Chai, G., Liu, Y., Li, X., Li, Z., Fan, Y., Zhang, Z.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Induced myeloid leukemia cell differentiation protein Mcl-1 | 157 | Homo sapiens | Mutation(s): 0 Gene Names: MCL1, BCL2L3 | 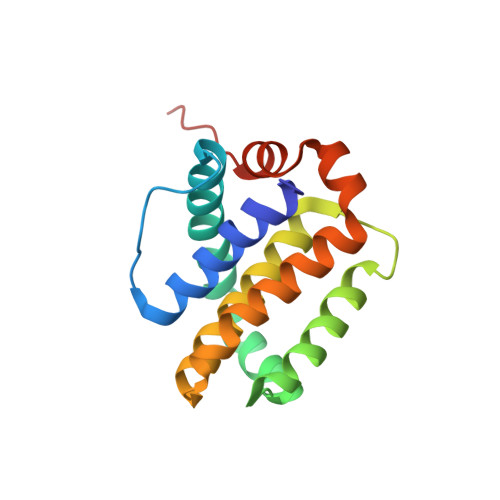 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q07820 (Homo sapiens) Explore Q07820 Go to UniProtKB: Q07820 | |||||
PHAROS: Q07820 GTEx: ENSG00000143384 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q07820 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mule BH3 peptide from E3 ubiquitin-protein ligase HUWE1 | 26 | Homo sapiens | Mutation(s): 0 | 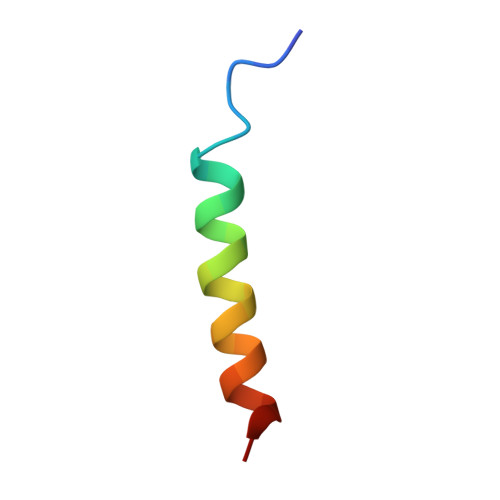 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q7Z6Z7 (Homo sapiens) Explore Q7Z6Z7 Go to UniProtKB: Q7Z6Z7 | |||||
PHAROS: Q7Z6Z7 GTEx: ENSG00000086758 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q7Z6Z7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 31.79 | α = 90.12 |
| b = 114.24 | β = 92.32 |
| c = 135.98 | γ = 90.01 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data collection |
| PHENIX | model building |
| Coot | data processing |
| XDS | data reduction |
| Coot | phasing |
| HKL-2000 | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China | China | 21402022 |
| National Natural Science Foundation of China | China | 21372036 |