Structure of Saccharomyces cerevisiae Rtr1 reveals an active site for an atypical phosphatase.
Irani, S., Yogesha, S.D., Mayfield, J., Zhang, M., Zhang, Y., Matthews, W.L., Nie, G., Prescott, N.A., Zhang, Y.J.(2016) Sci Signal 9: ra24-ra24
- PubMed: 26933063
- DOI: https://doi.org/10.1126/scisignal.aad4805
- Primary Citation of Related Structures:
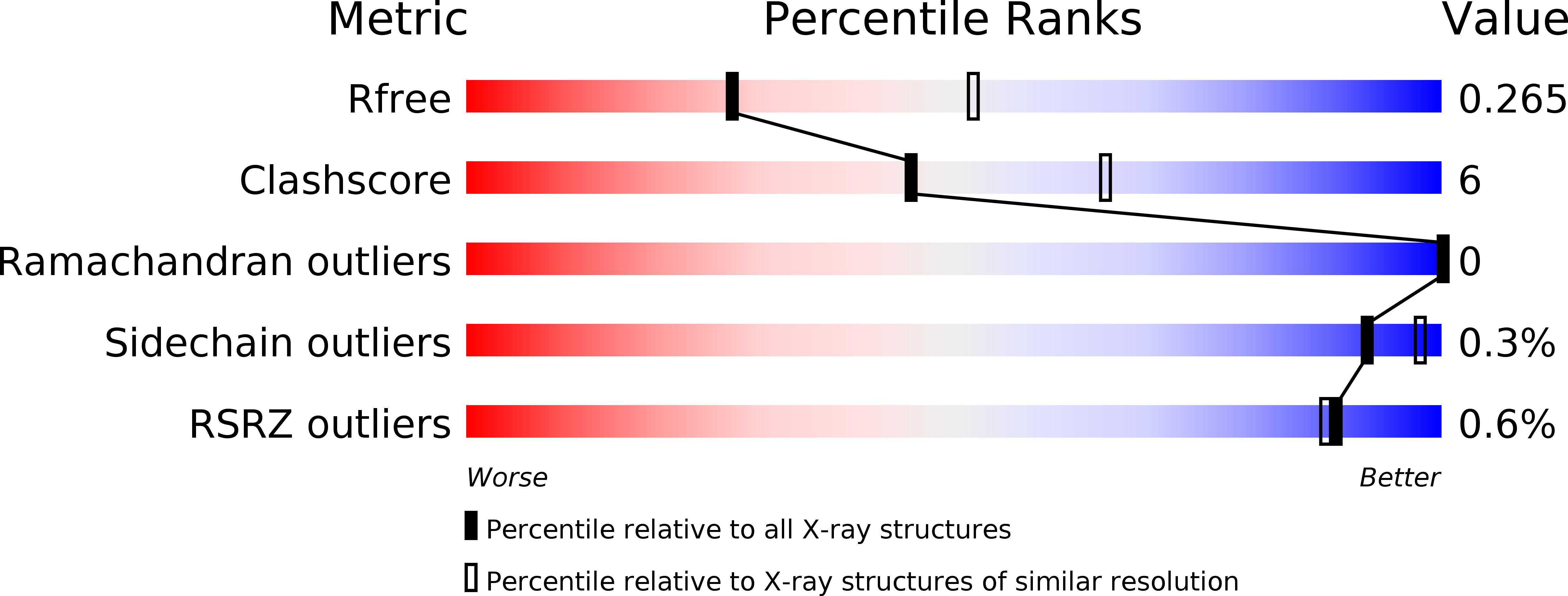
5C2Y - PubMed Abstract:
Changes in the phosphorylation status of the carboxyl-terminal domain (CTD) of RNA polymerase II (RNAPII) correlate with the process of eukaryotic transcription. The yeast protein regulator of transcription 1 (Rtr1) and the human homolog RNAPII-associated protein 2 (RPAP2) may function as CTD phosphatases; however, crystal structures of Kluyveromyces lactis Rtr1 lack a consensus active site. We identified a phosphoryl transfer domain in Saccharomyces cerevisiae Rtr1 by obtaining and characterizing a 2.6 Å resolution crystal structure. We identified a putative substrate-binding pocket in a deep groove between the zinc finger domain and a pair of helices that contained a trapped sulfate ion. Because sulfate mimics the chemistry of a phosphate group, this structural data suggested that this groove represents the phosphoryl transfer active site. Mutagenesis of the residues lining this groove disrupted catalytic activity of the enzyme assayed in vitro with a fluorescent chemical substrate, and expression of the mutated Rtr1 failed to rescue growth of yeast lacking Rtr1. Characterization of the phosphatase activity of RPAP2 and a mutant of the conserved putative catalytic site in the same chemical assay indicated a conserved reaction mechanism. Our data indicated that the structure of the phosphoryl transfer domain and reaction mechanism for the phosphoryl transfer activity of Rtr1 is distinct from those of other phosphatase families.
Organizational Affiliation:
Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX 78712, USA.