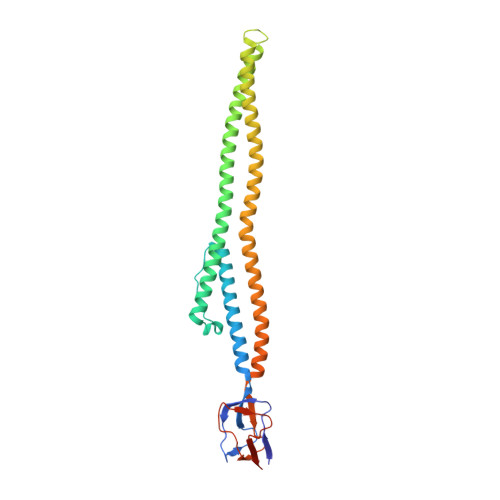
Crystal Structure of a Soluble Fragment of the Membrane Fusion Protein HlyD in a Type I Secretion System of Gram-Negative Bacteria
Kim, J.S., Song, S., Lee, M., Lee, S., Lee, K., Ha, N.C.(2016) Structure 24: 477-485
- PubMed: 26833388
- DOI: https://doi.org/10.1016/j.str.2015.12.012
- Primary Citation of Related Structures:
5C21, 5C22 - PubMed Abstract:
The protein toxin HlyA of Escherichia coli is exported without a periplasmic intermediate by the type I secretion system (T1SS). The T1SS is composed of an inner membrane ABC transporter HlyB, an outer-membrane channel protein TolC, and a membrane fusion protein HlyD. However, the assembly of the T1SS remains to be elucidated. In this study, we determine the crystal structure of a part of the C-terminal periplasmic domain of HlyD. The long α-helical domain consisting of three α helices and a lipoyl domain was identified in the crystal structure. Based on the HlyD structure, we modeled the hexameric assembly of HlyD with a long α-helical barrel, which formed a complex with TolC in an intermeshing cogwheel-to-cogwheel manner, as observed in tripartite RND-type drug efflux pumps. These observations provide a structural blueprint for understanding the type I secretion system in pathogenic Gram-negative bacteria.
Organizational Affiliation:
Department of Agricultural Biotechnology, Center for Food Safety and Toxicology, Center for Food and Bioconvergence, Research Institute for Agricultural and Life Sciences, Seoul National University, Room #200-1041, 1 Gwanak-ro, Gwanak-gu, Seoul 151-921, Republic of Korea.