Ligand efficient tetrahydro-pyrazolopyridines as inhibitors of ERK2 kinase.
Bagdanoff, J.T., Jain, R., Han, W., Poon, D., Lee, P.S., Bellamacina, C., Lindvall, M.(2015) Bioorg Med Chem Lett 25: 3626-3629
- PubMed: 26144345
- DOI: https://doi.org/10.1016/j.bmcl.2015.06.063
- Primary Citation of Related Structures:
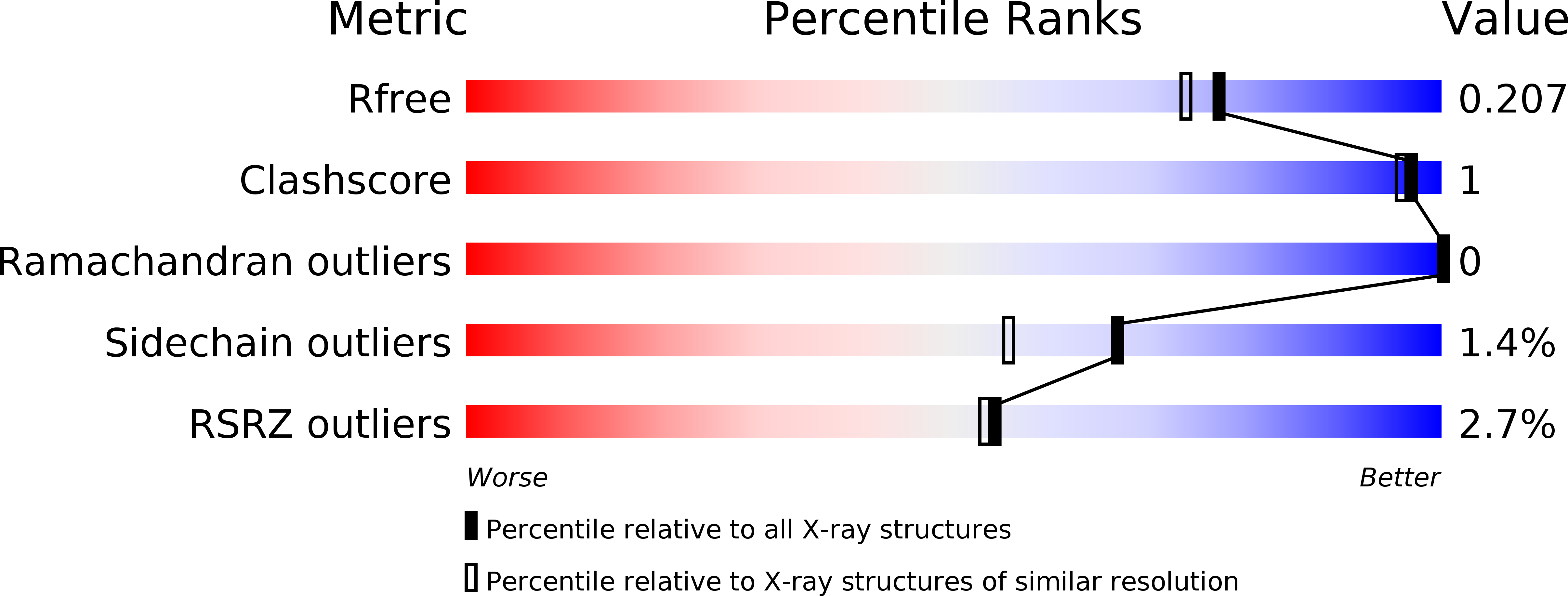
5BUE, 5BUI, 5BUJ - PubMed Abstract:
A series of structure based drug design hypotheses and focused screening efforts drove improvements in the potency and lipophilic efficiency of tetrahydro-pyrazolopyridine based ERK2 inhibitors. Elaboration of a fragment chemical lead established a new lipophilic aryl-Tyr interaction resulting in a substantial potency improvement. Subsequent cleavage of the lipophilic moiety led to reconfiguration of the ligand bound binding cleft. The reconfiguration established a polar contact between a newly liberated N-H and a vicinal Asp, resulting in further improvements in lipophilic efficiency and in vitro clearance.
Organizational Affiliation:
Global Discovery Chemistry/Oncology and Exploratory Chemistry, Novartis Institutes for BioMedical Research, 250 Massachusetts Ave., Cambridge, MA 02139, USA. Electronic address: jeffrey.bagdanoff@novartis.com.