Distinct Roles for the N- and C-terminal Regions of M-Sec in Plasma Membrane Deformation during Tunneling Nanotube Formation.
Kimura, S., Yamashita, M., Yamakami-Kimura, M., Sato, Y., Yamagata, A., Kobashigawa, Y., Inagaki, F., Amada, T., Hase, K., Iwanaga, T., Ohno, H., Fukai, S.(2016) Sci Rep 6: 33548-33548
- PubMed: 27629377
- DOI: https://doi.org/10.1038/srep33548
- Primary Citation of Related Structures:
5B86 - PubMed Abstract:
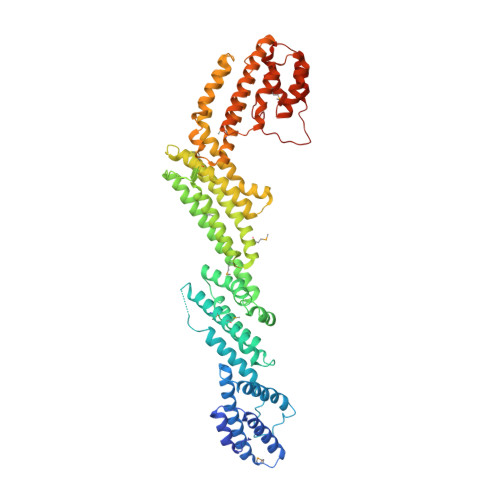
The tunneling nanotube (TNT) is a structure used for intercellular communication, and is a thin membrane protrusion mediating transport of various signaling molecules and cellular components. M-Sec has potent membrane deformation ability and induces TNT formation in cooperation with the Ral/exocyst complex. Here, we show that the N-terminal polybasic region of M-Sec directly binds phosphatidylinositol (4,5)-bisphosphate for its localization to the plasma membrane during the initial stage of TNT formation. We further report a crystal structure of M-Sec, which consists of helix bundles arranged in a straight rod-like shape, similar to the membrane tethering complex subunits. A positively charged surface in the C-terminal domains is required for M-Sec interaction with active RalA to extend the plasma membrane protrusions. Our results suggest that the membrane-associated M-Sec recruits active RalA, which directs the exocyst complex to form TNTs.
Organizational Affiliation:
Laboratory of Histology and Cytology, Graduate School of Medicine, Hokkaido University, North 15, West 7, Kita-ku, Sapporo 060-8638, Japan.