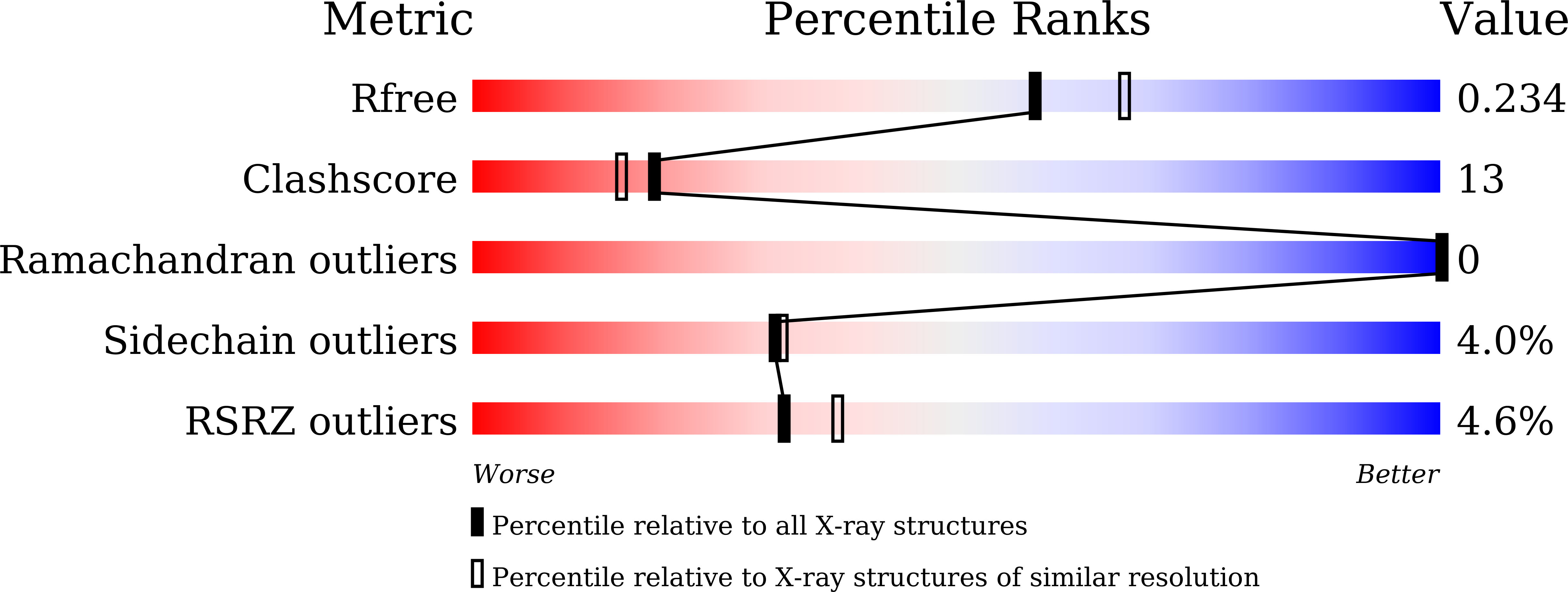
Catalytic properties and crystal structure of thermostable NAD(P)H-dependent carbonyl reductase from the hyperthermophilic archaeon Aeropyrum pernix K1.
Fukuda, Y., Sakuraba, H., Araki, T., Ohshima, T., Yoneda, K.(2016) Enzyme Microb Technol 91: 17-25
- PubMed: 27444325
- DOI: https://doi.org/10.1016/j.enzmictec.2016.05.008
- Primary Citation of Related Structures:
5B1Y - PubMed Abstract:
A gene encoding NAD(P)H-dependent carbonyl reductase (CR) from the hyperthermophilic archaeon Aeropyrum pernix K1 was overexpressed in Escherichia coli. Its product was effectively purified and characterized. The expressed enzyme was the most thermostable CR found to date; the activity remained at approximately 75% of its activity after incubation for 10min up to 90°C. In addition, A. pernix CR exhibited high stability at a wider range of pH values and longer periods of storage compared with CRs previously identified from other sources. A. pernix CR catalyzed the reduction of various carbonyl compounds including ethyl 4-chloro-3-oxobutanoate and 9,10-phenanthrenequinone, similar to the CR from thyroidectomized (Tx) chicken fatty liver. However, A. pernix CR exhibited significantly higher Km values against several substrates than Tx chicken fatty liver CR. The three-dimensional structure of A. pernix CR was determined using the molecular replacement method at a resolution of 2.09Å, in the presence of NADPH. The overall fold of A. pernix CR showed moderate similarity to that of Tx chicken fatty liver CR; however, A. pernix CR had no active-site lid unlike Tx chicken fatty liver CR. Consequently, the active-site cavity in the A. pernix CR was much more solvent-accessible than that in Tx chicken fatty liver CR. This structural feature may be responsible for the enzyme's lower affinity for several substrates and NADPH. The factors contributing to the much higher thermostability of A. pernix CR were analyzed by comparing its structure with that of Tx chicken fatty liver CR. This comparison showed that extensive formation of the intrasubunit ion pair networks, and the presence of the strong intersubunit interaction, is likely responsible for A. pernix CR thermostability. Site-directed mutagenesis showed that Glu99 plays a major role in the intersubunit interaction. This is the first report regarding the characteristics and three-dimensional structure of hyperthermophilic archaeal CR.
Organizational Affiliation:
Department of Bioscience, School of Agriculture, Tokai University, Aso, Kumamoto 869-1404, Japan.