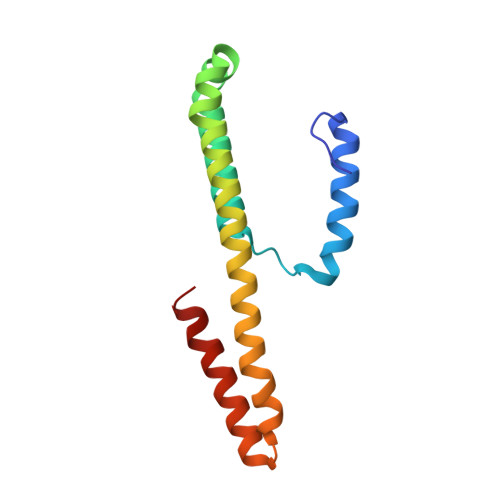
Structural Basis for Dimer Formation of the Crispr-Associated Protein Csm2 of Thermotoga Maritima.
Gallo, G., Augusto, G., Rangel, G., Zelanis, A., Mori, M.A., Campos, C.B., Wurtele, M.(2016) FEBS J 283: 694
- PubMed: 26663887
- DOI: https://doi.org/10.1111/febs.13621
- Primary Citation of Related Structures:
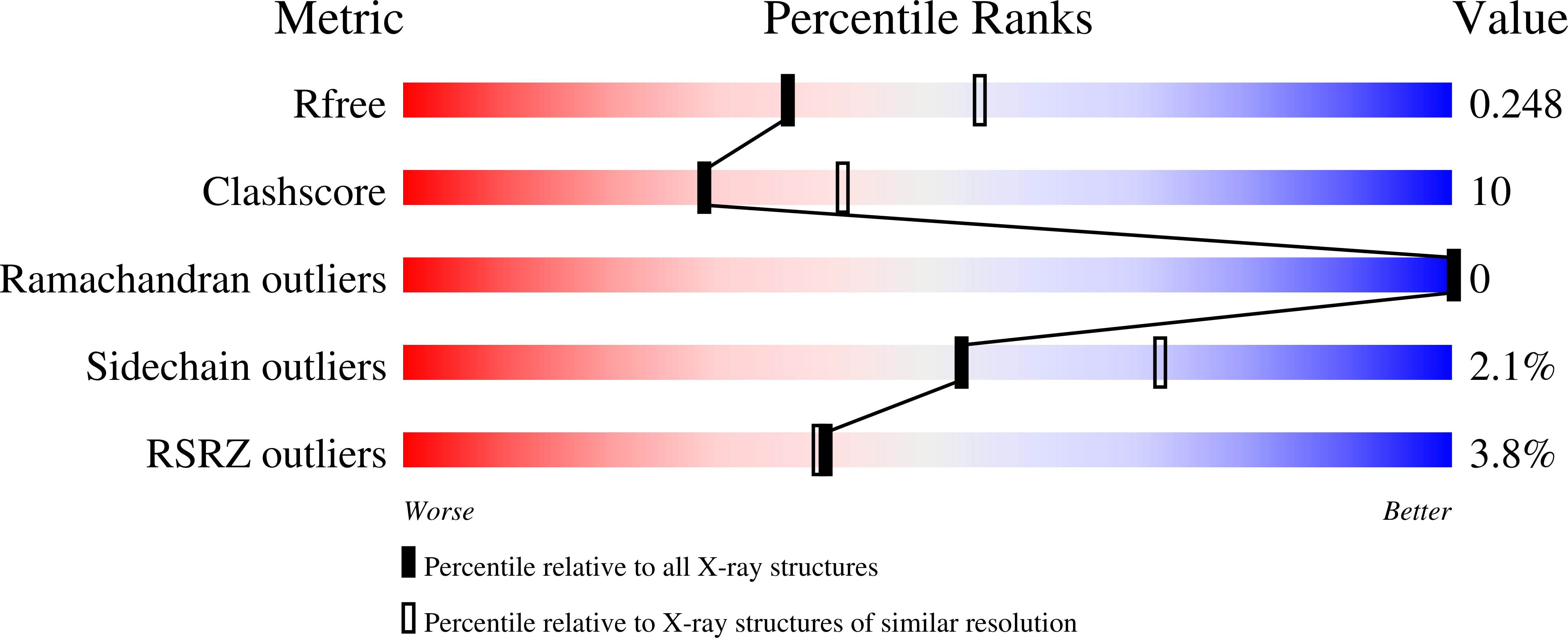
5AN6 - PubMed Abstract:
The clusters of regularly interspaced short palindromic repeats (CRISPR) and the Cas (CRISPR-associated) proteins form an adaptive immune system in bacteria and archaea that evolved as an RNA-guided interference mechanism to target and degrade foreign genetic elements. In the so-called type IIIA CRISPR-Cas systems, Cas proteins from the Csm family form a complex of RNPs that are involved in surveillance and targeting tasks. In the present study, we report the crystal structure of Thermotoga maritima Csm2. This protein is considered to assemble into the helically shaped Csm RNP complex in a site opposite to the CRISPR RNA binding backbone. Csm2 was solved via cadmium single wavelength anomalous diffraction phasing at 2.4 Å resolution. The structure reveals that Csm2 is composed of a large 42 amino-acid long α-helix flanked by three shorter α-helices. The structure also shows that the protein is capable of forming dimers mainly via an extensive contact surface conferred by its long α-helix. This interaction is further stabilized by the N-terminal helix, which is inserted into the C-terminal helical portion of the adjacent subunit. The dimerization of Csm2 was additionally confirmed by size exclusion chromatography of the pure recombinant protein followed by MS analysis of the eluted fractions. Because of its role in the assembly and functioning of the Csm CRISPR RNP complex, the crystal structure of Csm2 is of great importance for clarifying the mechanism of action of the subtype IIIA CRISPR-Cas system, as well as the similarities and diversities between the different CRISPR-Cas system.
Organizational Affiliation:
Department of Science and Technology, Federal University of São Paulo, São José dos Campos, Brazil.